Cell Biology
Explore Methods for Improving Resolution in Biological Microscopy
Analyzing cell structures and their organization is critical to understanding function within any cell type. However, visualizing many subcellular structures is impossible with traditional light microscopy due to the optical diffraction limit of ~200-300 nm.
Single-molecule localization microscopy (SMLM)—an efficient, straightforward process when performed with advanced microscope technology—solves this problem, enabling researchers to explore new cell biology frontiers in relating structure, organization, interaction, and stoichiometry to cellular function.
Microscopy Techniques for Cell Biology Imaging
Many interesting cellular structures are smaller than the optical diffraction limit of ~200-300 nm. These include the substructures of most organelles and all macromolecular machines, channels, and receptors.
Asking and answering questions about the molecular organization, interactions, and stoichiometry of these and other structures requires imaging of labeled structures at resolutions below the diffraction limit of light.
Conventional Microscopy Techniques in Cell Biology
Conventional light microscopy methods, like widefield and confocal microscopy, can image and identify specifically labeled cellular structures. However, they are limited in their ability to image below the optical diffraction limit. Alternatively, electron microscopy (EM) can achieve resolutions down to 0.1-0.2 nm laterally, but specific labeling or quantitative measurement of molecules within the cell is very challenging.
Super-Resolution Fluorescence Microscopy for Cell Biology
Super-resolution microscopy supports high-resolution imaging—down to 20 nm laterally and 50 nm along the optical axis —of specifically labeled cellular structures. It thereby bridges the gap between conventional light and electron microscopy (EM) techniques.
Mitochondria imaged with conventional fluorescence microscopy (left) vs. SMLM super-resolution microscopy (right) *Right: Mitochondria stained for TOM20 with Alexa 647, imaged with SMLM (dSTORM).
SMLM: See Biology in Nanoscale Detail
The high resolution (20 nm laterally) paired with the ability to image specifically labeled structures makes SMLM a powerhouse solution in cell biology research. Discoveries made with SMLM include the visualization of local movements of chromatin domains in HeLa cells, the eightfold symmetry within the nuclear pore complex of Xenopus oocytes and the radial ninefold symmetry of the centriolar protein CEP164, and the organization of connected tubular structures within the endoplasmic reticulum[1, 2].
Examples of Nanoscale Cellular Structures Resolved by SMLM
Super-resolution microscopy provides high-resolution imaging—down to 20 nm laterally and 50 nm along the optical axis—of specifically labeled cellular structures, bridging the gap between conventional light and electron microscopy (EM) techniques.
The resolution needed to visualize structures of interest must be taken into consideration when deciding if SMLM is the right solution for your sample. Some examples of the sizes of diffraction-limited structures better resolved by SMLM are provided. Examples* include (but are not limited to):
- Diffusion channel of the nuclear pore complex (40-50nm in length)
- Mitochondrial Nucleoids (~100 nm in diameter)
- Microtubules (24 nm in thickness)
- Ribosomes (~20-30 nm in diameter)
- Cylindrical pyrenoid tubules in the chloroplast (107 ± 26 nm in diameter)
- Endocytic pits in mouse cells (86±2.4nm in diameter)
- Tubular segments of cristae in rat liver mitochondria (30-40 nm in diameter)
- Plasma membrane caveolae (50-100 nm in diameter)
- Hook of the bacterial flagellum (~55 nm in length)
- Endoplasmic reticulum tubules and sheets (60-100nm diameter or thickness)
*Examples of nanoscale substructures and their dimensions are from 'The database of Useful Biological Numbers'. Link: https://bionumbers.hms.harvard.edu/search.aspx
Even if the structure you study is substantially larger than the resolution limit of PALM, dSTORM or PAINT, higher resolution will allow you to divide your structure into multiple spots, revealing otherwise invisible variation in biomolecule distribution, density, and location. Here are the number of resolvable units (we’re calling spots) in some common structures you may be interested in:
- The Mitochondrion or Lysosomes (~500nm in diameter or 25 spots)
- HIV-1 (~120nm in diameter or 6 spots)
- Yeast nucleus (~2 microns or 200 spots)
- Mammalian Nucleolus (~1 micron or 100 spots)
- Diameter of the Axon (300nm and up, or at least 15 spots)
Of the super-resolution microscopy approaches, Single-Molecule Localization Microscopy (SMLM) is arguably the best-suited for cell biology research.
Specifically, SMLM is the best method for studying specific nanoscale structures within cells when the research question requires:
- Visualization of molecules located down to 20 nm apart;
- Labeling of multiple specific molecular structures; or
- Quantitative data collection at this scale.
Applications of SMLM in Cell Biology
SMLM is already being used to answer previously unexplored questions in cell biology. SMLM is ideal for imaging multiple specific structures in 3D and at high resolution, as well as quantifying data. See the sample data and descriptions, below, to learn more about research applications that can be uniquely achieved with SMLM:
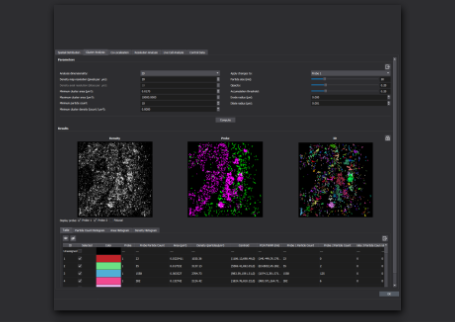
Molecular Quantification
With the top-hat illumination feature of Bruker's Vutara VXL, the whole field of view is illuminated evenly, resulting in uniform and reliable data acquisition across the entire region of interest. The resulting uniform blinking rates and intensities improves confidence in even detection performance across the whole field of view.
Each localized point contains statistical information that can be used for quantitative analysis, above and beyond its position. With SMLM, both relative and even absolute quantification of molecules is possible.
For example, a gap junction protein, connexin43, and a voltage-gated sodium channel, Nav1.5) are both located in intercalated disks; however, SMLM revealed that Cx43 and Nav1.5 are not expressed in equal quantity and do not form similar numbers of clusters[3].
Molecular Distribution
Whole field illumination is important for analyzing molecular distribution within a sample. The ability to uniformly illuminate the entire field of view, as well as the acquisition of statistical information associated with each localization, supports unbiased and quantitative analysis of the distribution of molecules throughout a sample.
As an example of molecular distribution analysis, SMLM allowed for the visualization of the distribution of the ParA ATPase with the ParB DNA binding protein localized to the cell poles, for the coordination of chromosome segregation and cell division[1].
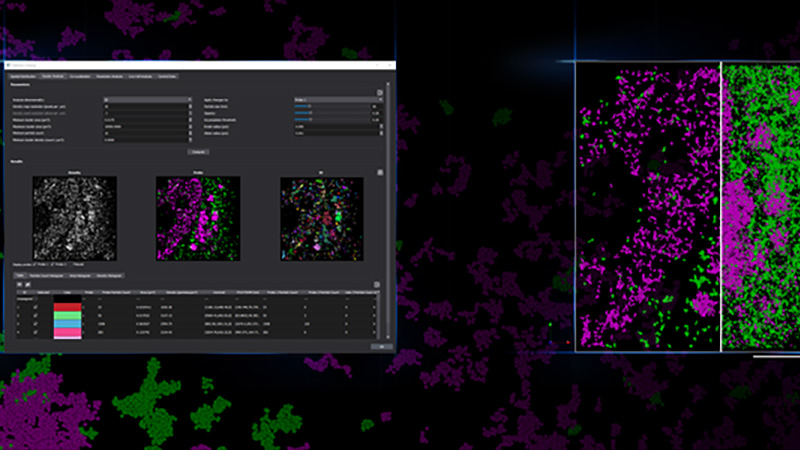
Colocalization
The goal of most co-localization experiments is to show the interaction between two or more biomolecules, or structures. Biologically meaningful interactions occur across space scales much smaller than the 200-300nm resolution limit of optical microscopy. With SMLM, one can image at a resolution of ~20 nm or less, allowing much more precise information about interactions between biomolecules. Traditionally direct, protein-protein interaction is demonstrated through techniques like Western Blot, while subcellular location, abundance and biological function can be demonstrated via microscopy. With single molecule localization, we can show fine-scale protein-protein interaction in situ within its relevant biological context within cells and tissues.
For example with SMLM, it was discovered that while there is a close association between connexin43 and Nav1.5, less than 20% of connexin43 clusters and only 10% of Nav1.5 clusters directly overlap with each other, and where they did overlap, it was minimal, which suggests that the clusters overlapped tangentially rather than representing a fully colocalized population of the two proteins[3].
Sample Images and Videos
Motion Models and Particle Tracking
With SMLM, the movement of molecules within a cell can be imaged in real-time with high spatial resolution. A successful example of single-particle tracking in a bacterial cell with SMLM showed that the actin homologue, MreB, moves in a circumferential pattern around the bacterial cell, driven by cell wall synthesis[4].
Right: Two experiments monitoring mitochondrial dynamics, labeled with (1) an orange HaloTag® dye (549) and (2) a photactivatable far-red dye (PA-JF-646®).
Fixed Sample Imaging
Microtubules acquired on the VXL - dSTORM images of fixed microtubules labeled with Alexa 647 using primary/seconday antibody labeling.
Live Cell Imaging
Acquired on the VXL - Imaging of live BSC1 cells labeled with Alexa 647 transferrin.
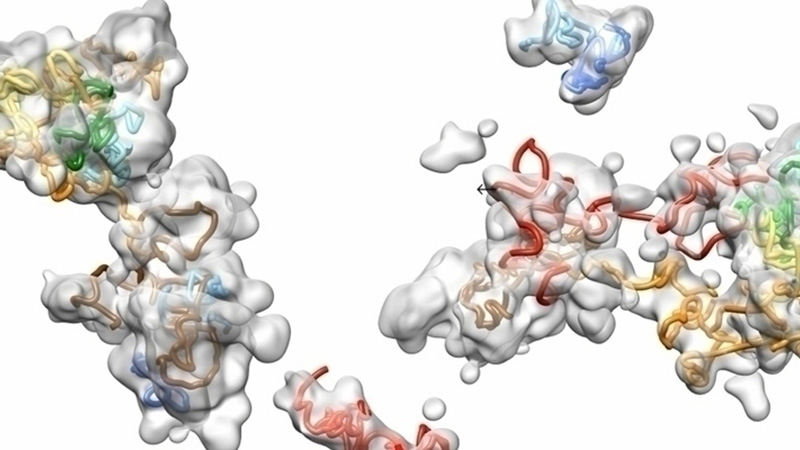
Point-Cloud Representation of Sodium Channels and N-cadherin Molecules
Image courtesy of:
Rengasayee Veeraraghavan, Ph.D. and Heather L. Struckman, B.S., M.S. at the Ohio State University Nanocardiology Lab
Complete Solutions for Single-Molecule Localization Microscopy
Bruker's Vutara VXL offers best-in-class ease of use and imaging depth. Key features supporting the needs of cell biology researchers include:
- Automated Workflows: SRX software makes it easy to acquire the necessary data sets, and workflows allow the user to focus on the biological question instead of tinkering with complicated microscope settings.
- Superior 3D Imaging and Sample Flexibility: Proprietary bi-plane detection provides three-dimensional information and enables imaging within tissue slices.
- Unlimited Multiplexing: The integrated Microfluidics Unit allows sequential labeling of an unlimited number of fluorescent probes.
- Expert Applications Support: Vutara VXL users can receive personalized guidance for sample prep optimization specific to their research objectives.
FAQ
Bruker's Vutara VXL is well-suited for cell biology research needs that require (1) high resolution, (2) 3D visualization, and/or (3) molecule-specific targeting and is applicable for samples ranging from single cells to tissue samples up to 50 microns thick.
The workflow and use of the Vutara VXL, microfluidics, and SRX software are streamlined and Bruker scientists are available to provide personalized support—from sample preparation to data analysis—to Vutara VXL users.
SMLM achieves super-resolution by localizing single, blinking dye molecules with high precision. Diffraction-limited light microscopy can't distinguish two dye molecules within a distance less than 300 because their point-spread functions overlap too much. In SMLM, only one of the two dye molecules is active, while the other one is dark. Now, we can determine the position of this molecule with high precision. After a while, the active dye is rendered dark, and the previously dark dye becomes active. Now we can determine the position of the second molecule with high precision. The on-off switching of the dyes can happen actively (e.g., photo-activatable fluorescence proteins in PALM) or spontaneously (e.g., Alexa Fluor 647 in dSTORM).
A broad range of sample types can be imaged with SMLM. Both fixed and live samples can be imaged with SMLM, although fixed-sample imaging is more common. With the Vutara VXL, one can image a variety of sample types, ranging from cell culture and tissue slices, whole organisms, such as Drosophila larvae and C. elegans, and hydrogels up to 100 microns thick.
With the bi-plane technology of the Vutara VXL, one can image up to 100 microns from the coverslip in hydrogels, and up to 50 microns for thick samples, such as tissue slices or even whole organisms. Tissue clearing may be required for thick samples of tissue or whole organisms for optimal imaging.
Although SMLM is a new and advanced imaging approach, sample preparations and imaging protocols are well understood and documented. Bruker applications specialists can help users to choose appropriate labeling strategies and imaging settings.
Although less common than fixed-sample imaging, live-sample imaging can be performed with SMLM. View this webinar discussing the preparation of live samples for imaging. See "References and Resources" for more information on live-cell imaging techniques and best practices[3,4].
References
- [1] Sigal, Y.M., Zhou, R. and Zhuang, X., 2018. Visualizing and discovering cellular structures with super-resolution microscopy. Science, 361(6405), pp.880-887.
- [2] Schermelleh, L., Ferrand, A., Huser, T., Eggeling, C., Sauer, M., Biehlmaier, O. and Drummen, G.P., 2019. Super-resolution microscopy demystified. Nature cell biology, 21(1), pp.72-84.
- [3] Veeraraghavan, R. and Gourdie, R.G., 2016. Stochastic optical reconstruction microscopy–based relative localization analysis (STORM-RLA) for quantitative nanoscale assessment of spatial protein organization. Molecular biology of the cell, 27(22), pp.3583-3590.
- [4] Gahlmann, A. and Moerner, W.E., 2014. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nature Reviews Microbiology, 12(1), pp.9-22.
- [5] Frigault, M.M., Lacoste, J., Swift, J.L. and Brown, C.M., 2009. Live-cell microscopy–tips and tools. Journal of cell science, 122(6), pp.753-767.
- [6] Nizami, Z.F., 2010. Live Cell Imaging, A Laboratory Manual, Robert D. Goldman, Jason R. Swedlow, and David L. Spector (Eds.). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 2010. ISBN 978-0-87969-893-5. Microscopy and Microanalysis, 16(5), pp.649-649.
- [7] Manley, S., Gillette, J.M. and Lippincott-Schwartz, J., 2010. Single-particle tracking photoactivated localization microscopy for mapping single-molecule dynamics. Methods in enzymology, 475, pp.109-120.
- [8] Martin-Fernandez, M.L. and Clarke, D.T., 2012. Single molecule fluorescence detection and tracking in mammalian cells: the state-of-the-art and future perspectives. International journal of molecular sciences, 13(11), pp.14742-14765.
- [1] Sigal, Y.M., Zhou, R. and Zhuang, X., 2018. Visualizing and discovering cellular structures with super-resolution microscopy. Science, 361(6405), pp.880-887.
- [2] Schermelleh, L., Ferrand, A., Huser, T., Eggeling, C., Sauer, M., Biehlmaier, O. and Drummen, G.P., 2019. Super-resolution microscopy demystified. Nature cell biology, 21(1), pp.72-84.
- [3] Veeraraghavan, R. and Gourdie, R.G., 2016. Stochastic optical reconstruction microscopy–based relative localization analysis (STORM-RLA) for quantitative nanoscale assessment of spatial protein organization. Molecular biology of the cell, 27(22), pp.3583-3590.
- [4] Gahlmann, A. and Moerner, W.E., 2014. Exploring bacterial cell biology with single-molecule tracking and super-resolution imaging. Nature Reviews Microbiology, 12(1), pp.9-22.
- [5] Frigault, M.M., Lacoste, J., Swift, J.L. and Brown, C.M., 2009. Live-cell microscopy–tips and tools. Journal of cell science, 122(6), pp.753-767.
- [6] Nizami, Z.F., 2010. Live Cell Imaging, A Laboratory Manual, Robert D. Goldman, Jason R. Swedlow, and David L. Spector (Eds.). Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY; 2010. ISBN 978-0-87969-893-5. Microscopy and Microanalysis, 16(5), pp.649-649.
- [7] Manley, S., Gillette, J.M. and Lippincott-Schwartz, J., 2010. Single-particle tracking photoactivated localization microscopy for mapping single-molecule dynamics. Methods in enzymology, 475, pp.109-120.
- [8] Martin-Fernandez, M.L. and Clarke, D.T., 2012. Single molecule fluorescence detection and tracking in mammalian cells: the state-of-the-art and future perspectives. International journal of molecular sciences, 13(11), pp.14742-14765.