Tech Note: Single Molecule Force Spectroscopy with the ForceRobot® 300
Single molecule force spectroscopy (SMFS) enables the investigation of both the internal properties of single molecules and the binding and interactions between molecules. This technique places particular demands on the instrument — beyond the standard requirements for high-quality atomic force microscopy imaging — because of the demands of working with individual molecules. To meet the unique challenges of SMFS experimentation, the ForceRobot® 300 was developed as a dedicated force spectroscope.
This technical note provides a detailed examination of the unique features and capabilities of the ForceRobot® 300 system and how it overcomes the most significant challenges of SMFS experiments.
Readers can expect to learn about:
- Specific instrument requirements and other practical considerations for SMFS experiments
- The unique capabilities and advantages of using the ForceRobot® 300 for quantitative SMFS
- How the ForceRobot® 300’s unique mechanical design and automation features achieve higher stability, accuracy, and throughput than other measurement systems
Introduction
Single molecule force spectroscopy with the ForceRobot® 300
The atomic force microscope (AFM) allows imaging or manipulation of materials ranging from individual molecules such as DNA and proteins to whole cells. The sensitive force detection and control have naturally led to a range of experiments where single molecules are investigated without lateral scanning or imaging. These force spectroscopy experiments probe the response of the molecule over a spectrum of forces. Single molecule force spectroscopy enables the investigation of internal molecular properties such as backbone elasticity and length or intramolecular binding, for example protein unfolding. The binding and interactions between molecules can also be probed, as intermolecular binding, for instance ligand-receptor or DNA hybridization [1]. It can even be possible to differentiate ligand and inhibitor binding to membrane proteins on the basis of their unfolding signatures [2]. By varying the experimental parameters, the kinetics and energy landscape of the binding can also be measured [3].
Single molecule force spectroscopy (SMFS) places particular demands on the instrument, leading to the development of the ForceRobot®300 as a dedicated force spectroscope. The requirements partly overlap with those of a high-quality atomic force microscope, such as highprecision piezo elements and sensitive, low-noise detection systems. The positioning and deflection detection of the cantilever at sub-nanometer precision are a prerequisite for high-quality force experiments. Other requirements are specific to the force spectroscopy experiments, because of the demands of working with individual molecules.
SEE THE LATEST GENERATION FORCEROBOT:
There are two important aspects to single molecule experiments when the molecules are investigated sequentially. The first is that many repeats of the experiment will be required to build up a picture of the ensemble behavior. The binding or unbinding of interactions close to the thermal energy is strongly influenced by the instantaneous state of a particular molecule, requiring multiple measurements to build up a histogram of binding or unbinding forces. The advantage of single molecule experiments is that within the ensemble, it is possible to classify different responses of different molecules, for example different unbinding pathways.
The second aspect comes from the practical questions of how to arrange a single molecule at the tip of the cantilever, and how to know there is only one molecule. The general solution is to adjust the surface coverage until only around one in ten force curves show a specific response. Then the chances of collecting data from two or more molecules interacting becomes small enough to be able to interpret the force curves properly. So most of the force curves will not show the specific interaction that is being investigated, and at the same time many of these specific events must be measured in order to build up a full picture. To make this process efficient, a dedicated instrument is required which can collect thousands of force curves automatically without user input or adjustment. The data analysis should also be automatic, to filter out uninteresting force curves, and apply the particular fitting or other routines to analyze the interesting curves.
The ForceRobot®300 provides a turn-key instrument dedicated to quantitative single molecule force spectroscopy. The automated setup is designed for longterm unattended measurements, and the recording and processing of large amounts of data. In addition, optical integration is included, to enable location of bound protein based on fluorescence. This allows targeted measurements where the molecules are located. The ForceRobot®300 delivers the stability, reproducibility, automation, throughput and environmental control required to efficiently probe single molecules.
Stability and drift compensation for longterm measurements
At the heart of force spectroscopy measurements is the requirement to collect a large number of reproducible data sets. Obviously the calibration and accuracy of the system is critical to measuring the highest quality data. Beyond this, the long term stability is vital to enable long experiments without constant user attention. The AFM cantilevers used for the force spectroscopy experiments bend over time in reaction to temperature changes, protein adsorption and many other effects. To overcome this, the ForceRobot® 300 has an automated correction to readjust the beam deflection system between force curve cycles.
The initial alignment of the system takes place completely automatically, after selection of the chosen cantilever position. The laser spot and cantilever can be easily identified by using the CCD camera integrated in the ForceRobot® head. The motorized readjustment can be set to occur automatically as often as required during longer experiments. The camera can also be used to monitor the sample or find interesting locations.
High-quality data are vital for successful analysis. The lowest electronic noise floor and the most rigid mechanical design are essential. The highest accuracy and stability of the instrument is ensured by integrated capacitive position sensors with exceptional performance. For minimized drift, the system design is symmetric. For the detection of smallest variations in force curves, JPK improved the sensitivity and increased the detection bandwidth without compromising the noise levels. Utilizing a high sampling rate and a high number of data points per curve, the instruments offers a unique measurement performance. Together with a rigorously drift-minimized mechanical design, auto-calibration and the automated drift compensation enable long-term measurements with consistently high data quality.
High-throughput data generation
As highlighted in the introduction, large data sets need to be generated in force spectroscopy experiments in order to achieve meaningful results. The ForceRobot®300 overcomes the limitations of traditional force measurements by automating routines and providing intelligent software for experiment design. Tens of thousands of force curves are generated in a matter of hours in unattended mode, and force curves without events are automatically filtered before processing. The possibility to integrate the ForceRobot® into an inverted optical microscope enables fluorescence microscopy (e.g. epifluorescence, confocal, FRET) simultaneously with the force experiment, as shown in figure 2.
The ForceRobot® software package is characterized by its completely automated data acquisition but it is open enough to give the user full experimental control. The system comes with an intuitive user interface — a new way to structure the data acquisition and analysis. The ExperimentPlanner™ increases productivity with enhanced experimental flow control. The user can easily design experiments with temperature ramps, position movements, different force ramps or liquid exchange. These experiments can be saved and reloaded upon demand.
Hand in hand with this feature, the RampDesigner™ was developed to offer a new level of experimental flexibility. The user can freely combine segments of a force-distance cycle to customize advanced force curve procedures. Force ramps and clamps can be combined with length ramps and delays, allowing flexible composition of experimental parameters such as triggers, ramps and pulling speeds (see figure 3). Force clamp or refolding experiments are of particular interest for protein molecules [4,5]. For every segment, individual sample rates, gains and setpoints can be specified. The whole set of parameters for a particular force curve can be saved and reloaded, allowing reproducible measurements over many experiments. Different force ramps can also be combined using the ExperimentPlanner™ to build up complex experiments varying pulling speed or temperature.
Batch Filtering and Analysis
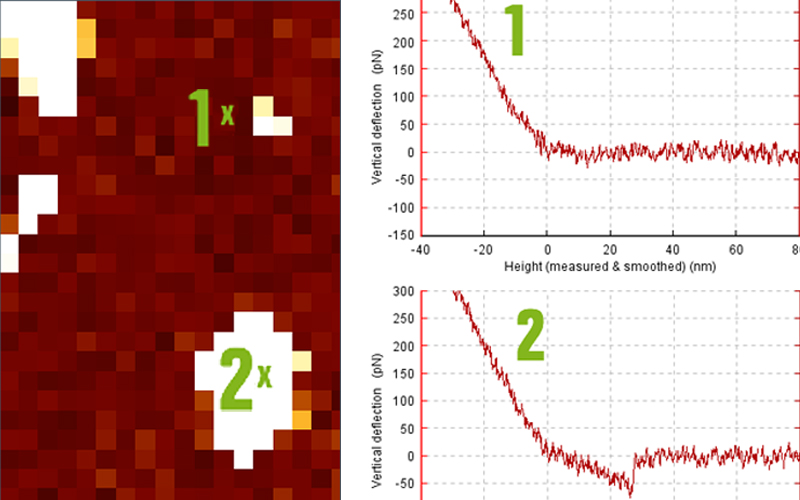
Once the ForceRobot runs in automated mode, it generates several thousand curves in the course of a few hours. In order to deal with this enormous amount of data, intelligent filtering and batch processing needs to be performed. The ForceRobot software allows for both online and offline filtering of successful force curves. Signal thresholds can be set for the recording of data in order to avoid filling disk space with force curves lacking events. An additional offline filtering can be performed that allows a force curve classification.
The settings for the batch processing in the JPK Data Processing software can also be saved for the individual processing steps. These include fitting procedures such as worm-like-chain (WLC), freely-jointed-chain (FJC) and step fitting algorithms. In figure 4, a result of SMFS experiments performed on the ForceRobot® 300 is presented. The curve shows the unfolding of filamin during a force extension experiment (ddFLN, samples kindly provided by Matthias Rief, TU Munich). Filamin is an actin cross-linking protein which consists of six tandem repeats that can be unfolded sequentially. Using the RampDesigner™ forces curves with the following segments were generated:
- Approach with a threshold of lnN at a speed of 1 pm/s
- Delay on the surface of 2 seconds
- Retraction with a length of 500nm at a speed of Ipm/s
By automatically and repeatedly rescanning a grid of spots, several hundred curves were generated in unattended mode in only one hour.
The curves have been filtered using a threshold of 50 PN and the peaks were fitted with the worm-like-chain model. The fit results include the persistence and the contour length of the protein, the loading rate and the unfolding force. After the automated fit of a batch of data, the results can be easily exported into statistics software, to plot histograms.
Environmental control and sample exchange
The mechanical properties of single molecules are strongly influenced by their environment, and the unique environmental control of the ForceRobot has been designed with this in mind. Heating and cooling is available over a wide range of temperatures. This enables the study of biological samples at physiologically relevant temperatures or polymer samples in phase transition. The JPK FluidicsModule™ allows the design and performance of experiments with different buffer composition, pH values or chemical agents. Up to eight different liquids, such as buffer solutions, can be connected to the fluid cell, offering a maximum flexibility to perform experiments under varying buffer conditions for one molecule. The flow rates and liquid choice are controlled from the software and included in the ExperimentPlanner™ settings.
Outlook
Single-molecule force spectroscopy can generate valuable new insights on molecular mechanics. With the ForceRobot®300, JPK offers a new tool for automated spectroscopy of force experiments on single molecules, such as proteins and polymers. The applications range from the unfolding of proteins to the probing of the kinetic landscape of protein-ligand interaction and the quantification of mechanical properties of biopolymers. Conventional SMFS can record force-distance curves in a continuous mode, but the user has to manually adjust the other parameters, such as compensation of drift, recalibration, temperature control and buffer adjustments. The ForceRobot® 300 offers a full automation of the experiments, including calibration, drift compensation, experiment design and recording in unattended mode. The setup offers the stability for long-term measurements, and the software is especially designed for automated recording and processing of high quantities of data. A large batch of data can be subject to a reproducible filtering and data analysis, which include fitting models like WLC, FJC, step fitting or elasticity modules.
The unique environmental control of the ForceRobot furthermore takes care of the experimental conditions. The setup is tailored to the fact that mechanical properties of single molecules are strongly influenced by their environment. Heating and cooling is available over a wide range of temperatures. The JPK FluidicsModule™ allows the design and performance of experiments with different buffer conditions.
Such an intelligent approach to screen molecular interactions allows to establishing SMFS as a standard tool to analyze the interacting mechanisms of biomolecules. In the end, the user can focus on the experimental results while the system delivers high quality data in unattended operation.
References
- Y.J. Jung, B.J. Hong, W. Zhang, S.J.B. Tendier, P.M. Williams, S. Allen and J.W. Park "Dendron Arrays for the Force-Based Detection of DNA Hybridization Events" J. Am. Chem. soc. (2007) 129: 9349-9355
- Kedrov, C. Ziegler and D.J. Muller "Differentiating Ligand and Inhibitor Interactions of a Single Antiporter" J. Mol. Biol. (2006) 362: 925-932
- Kedrov, M. Appel, H. Baumann, C. Ziegler and D.J. Muller "Examining the Dynamic Energy Landscape of an Antiporter upon Inhibitor Binding" J. Mol. Biol. (2008) 375: 1258-1266
- Oberhauser, P. Hansma, M. Carrion-Vazquez and J. Fernandez, "Stepwise unfolding of titin under forceclamp atomic force microscopy", PNAS (2001)98:468
- M. Rief, M. Gautel, F. Oesterhelt, J.M. Fernandez, H.E. Gaub "Reversible Unfolding of Individual Titin Immunoglobulin Domains by AFM" Science (1997) 276:1109-1112