Application Note: Dissecting T Cell Mechanosensing at Molecular and Cellular Scales: The Use of Nano-Force Tools
AFM can also be combined with fluorescence microscopy to characterize, in real-time, some of the intercellular signals that are generated when the proteins present on the cell surface are stimulated. When used in combination with optical tweezers (OT), a detailed characterization of cellular mechanics can be obtained. By varying the shape and size of the indenter over different scales, information on the sub-membrane organization of the cytoskeleton can also be obtained.
When AFM or OT are combined with fluorescence imaging, it could lead to the characterization of the transfer function of the membrane protein system. When AFM is used in combination with fluorescence microscopy, 3D functional structures on the cell surface can be revealed. Finally, combining AFM and OT allows the reproduction of environments more similar to physiological ones, as one can assess the interaction between a T cell and an antigen presenting cell using SCFS, and when a helper T cell is brought into contact, the modulation of the recognition forces can be recorded. Ultimately, the use of nano-force tools is full of promise for the emerging fields of immune-biophysics and immunomechanics.
Readers can expect to learn about:
- The evaluation of TCR recognition on living T cells at the single molecule scale;
- The characterization of substrates for interaction with T cells; and
- The characterization of model antigen-presenting cells
KEYWORDS: Optical Tweezers; NanoTracker 2; AFM; NanoWizard; OT-AFM; Cell Biology; T Cells; Membrane Tethers; Membrane Pulling; Mechanosensing; Nanoscale Force
Introduction
The key function of T lymphocytes during an immune response is to scan the surface of surrounding cells and detect, via the membrane T cell receptor (TCR), the presence of foreign peptide antigens on antigen presenting cells (APC) among the many self-peptides presented by the Major Histocompatibility Complexes (MHC). A TCR-peptide-MHC (pMHC) interaction is required for the activation of T-cells and subsequent actions, such as proliferation, which is the essence of the adaptive immune response. In addition, TCR-pMHC interactions constantly provide "survival signals" in order to maintain a steady population of memory cells, which constitute our long-term immunity.1
TCR dependent signaling must therefore be both rapid and sensitive in order to efficiently detect the presence of very low numbers of foreign peptide antigens, and at the same time filter out self-peptide/MHC generated «noise», so as not to harm healthy cells and normal tissue. The different ways TCR-peptide-MHC binding events are processed by the cellular signaling machinery of the lymphocytes remains a critically important question for both the development and the function of the adaptive immune response.
In particular, a lot of effort has been made to quantify:
- The kinetics of the TCR-peptide-MHC bonds;
- The number of interacting partners;
- The type and role of co-stimulatory molecules;
- The spatial organization of the activating molecules; and
- The contribution of other physical parameters, such as the forces exerted on the molecular bonds.2
For this reason, the cytoskeletal architecture of T cells appears to play an ever more central role in their recognition and activation properties, but this needs to be further clarified and quantified. It allows the T cells to exert forces on the APC, down to the single molecule scale. It has been proposed that these forces are a key factor in the capacity of T cells to selectively and sensitively recognize foreign peptides and in their activation. T cells can feel the overall rigidity of the substrate they are in contact with and, in addition to its biochemical properties, use it as a supplementary signal. It is thought that, to a certain extent, mechano-transduction plays a role in the activation of T cells, from the molecular scale to a more global cellular scale.3
EXPLORE THE BIOAFM APPLICATIONS GALLERY:
As a result, the aim of our investigations is to examine how the micro- and nano-organization of activation-related surface molecules affect or are affected by the micro-mechanical properties of T cells (such as recognition, adhesion, elasticity, membrane tension) using advanced biophysical techniques based on force application/measurement such as Atomic Force Microscopy (AFM) or Optical Tweezers (OT). These techniques allow the measurement of cell elasticity, viscosity and adhesion during signaling.
OT and nano-indentation by AFM allow the investigator to gain quantitative information about the elasticity and viscoelasticity of cells. Membrane tension can be measured by using AFM or OT to pull tethers from the membrane.
In the following, we will go into detail on how both AFM and OT can be important tools in the field of immuno-mechanics.
LEARN MORE:
Evaluation of TCR recognition at the single molecule scale, on living T cells
In a first attempt to understand which physical parameters are recognized by T cells when they come in contact with an antigen presenting cell, we designed a single molecule study to determine the probability of adhesion and the forces of detachment at the surface of living lymphocytes. We used SMFS (Single Molecule Force Spectroscopy) where the tips of soft AFM cantilevers were decorated with recombinant pMHC molecules (Figure 1).
A: Schematics of the experiments on living T cell hybridoma, with the methodology used to decorate AFM cantilevers with pMHC.
B: Typical single molecule separation event, showing the very low specific forces (~ 20 pN) that were recorded, to be compared to larger forces obtained for classical adhesion molecules. Adapted from.4
We used them to probe the mechanics of the T cell under very low forces and for very short periods of contact time (-100 msec). This resulted in infrequent recognition events. By varying the peptide load in the pocket of the MHC, we showed that the recognition was indeed specific, but that the rupture forces we observed under our experimental conditions were not peptide-dependent.4
It was later shown, via techniques using softer springs (a laser for OT; a Red Blood Cell for a biomembrane force probe) that indeed, the TCR can behave as a mechanosensory molecule. The forces that the T cell feel via the TCR are now thought to help recognition be highly specific and rapid.5
LEARN MORE:
Real-time combination of AFM and fluorescence microscopy
We performed AFM indentation of T cells simultaneously with intracellular Ca2+ fluorescence imaging to gain quantitative information on the forces at play during the different activation phases.6 We used an original internal timer signal and AFM based mechanical stimulation to apply a mechanical stimulation that can, in addition, be made specific via, e.g., anti CD3 antibodies. We have developed micro-manipulation techniques to decorate AFM cantilevers with beads of diameters ranging from 1 to 50 µm in order to control the shape and size of the stimulating surface. Stimulation can be performed with either continuous contact or with a succession of timed, short stimulations in order to dissect how the "signaling black box" answers the same specific signal, but with a specified spatial or temporal distribution. This approach, in a way, is a typical physicist method for dissecting the "transfer function" of an unknown system. Here, biology allows us to interfere with the "black box" thanks to the use of mutants or molecules that can impact specific parts of the signaling cascades or functions. We also developed protocols to record the modification of the mechanical properties of T cells with optogenetic tools, for example, photoactivatable Rae, a small GTP-ase protein.6
Characterization of substrates for interaction with T cells
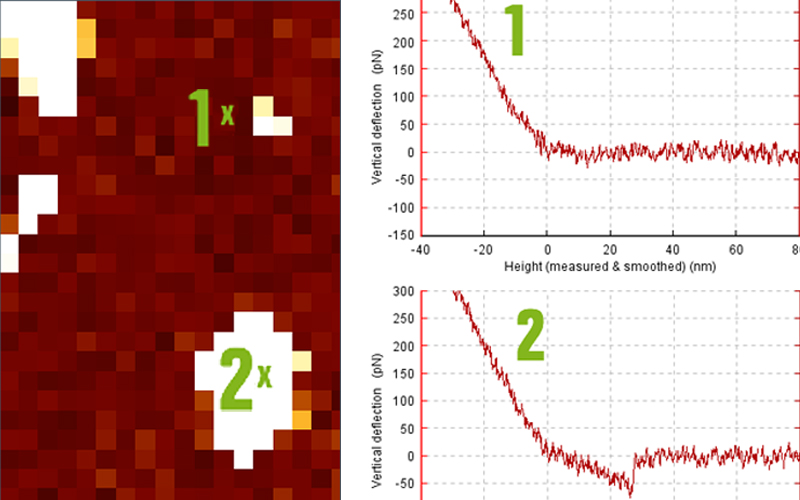
Two AFM modes can be used to characterize the topology or the mechanical properties of an artificial substrate which will interact with the T cells. Firstly, one can image the substrate at the nanoscale and observe its roughness or its surface structure, e.g. using micro-contact printing or nano-scale patterns.7, 8 Secondly, when preparing soft polymeric substrates or soft gels, substrate indentation or force mapping can be used to precisely characterize the mechanical properties of the substrate the cell is exposed to at the subcellular scale using a thin tip/small scan range, or at the cellular scale using a bead for cellular contact/ larger scan range. We observed that the mechanical properties of the substrate may lead to unconventional spreading behavior of the T cells depending on the molecular decoration present at the surface.3
LEARN MORE:
Characterization of model antigen presenting cells
We shaped model APCs (COS-7 cells) on stamped fibronectin, while preventing the adhesion and activation of the T cells around them.9 Micropatterns were used to pull on the edge of the cell to create a large lamellipodium that was thin enough to observe the early contacts of T cells through it using advanced surface microscopies such as TIRF (with membrane labelled T cells) or RICM (without any labelling).
This structure allows us to have a fully controlled, fluid, cellular environment, the composition of which is controllable by transfection and is closer to physiological conditions than textured substrates and supported lipid bilayer approaches.10 The 3D topography of the lamellipodium was characterized using different pattern sizes and a combination of TIRF, confocal (membrane, cytoskeleton), multi-color RICM reconstruction in conjunction with AFM imaging and force mapping.11 We also used classical AFM indentation to characterize the Young's modulus of different populations of COS cells expressing different receptors and molecules in regard to T cell mechanics (unpublished data, coherent with).12
T lymphocyte/antigen presenting cell forces
In order to simulate physiological situations more accurately, we approached the APC vs. T cell situation by attaching a model APC (a transfected COS-7 cell) to an AFM cantilever and brought it in contact with a (Ca2+ reporter loaded) T cell of known shape.9 This method, called Single Cell Force Spectroscopy (SCFS), allows the evaluation of the contact mechanics between cells, the evolution of the contact and the forces needed for separation, from single molecules to entire cell scales.14
It has recently been applied to T cell/APC contact over long contact times and after formation of the immune synapse,13 but only adhesion at the cellular scale was recorded and simultaneous observation of the early activation of the cells was not made.
A: Schematics of the experiments, where a calcium reporter loaded T cell, gently immobilized on a PLL coated surface, is brought into contact with an APC attached to an AFM lever.
B: Superposition of transmission and fluorescence images, with the APC (white arrowhead), the contacted T cell (green arrowhead) and a control/non contacted T cell (red arrowhead).
C: Force signal, presented with the fluorescence signal emitted by contacted and control T cells. The rise in calcium follows the contact, as detected by force rise, by - 1 min, which is coherent with literature.
Combining SCFS with our technique of simultaneous AFM/fluorescence to record the activation pattern before, during and after contact, the early physical determinants of T cell activation can be analyzed, such as which forces are required or created by T cells to integrate a biochemical or biomechanical signal (Figure 2). Depending on the contact times, the molecules involved and potential signaling required to reinforce the cell/cell adhesion, such approaches may need an extended piezo range (up to 100 µm or even more). It may also be necessary to coordinate the fluorescence-detecting lens with a supplementary piezo, all of which is possible with the Bruker CellHesion module, an add-on of the Bruker NanoWizard AFM.14 Cell/cell adhesion forces, numbers and separation distances are indeed larger compared to usual SCFS (cell vs. substrate) experiments, requiring adaption of the spring constant of the cantilever to be able to record the forces, and, importantly, the puling range, which is usually limited to 10-15µm in conventional AFMs.
Measuring T cell cortical tension by pulling membrane tethers
Using e.g. lectin decorated cantilevers, membrane 0 tubes or tethers can be pulled using AFM. Cortical cell tension can be estimated based on the force needed to extract them from the cell, and, using drug-induced perturbation of the cell cytoskeleton in a parallel experiment, the cell's membrane tension and the interaction energy between the membrane and the cytoskeleton can be evaluated.15, 16
Using Optical Tweezers (for us, the Bruker NanoTracker 2), the resolution in force can be increased compared to AFM, and such experiments can be performed in combination with fluorescence imaging of the membrane (Figure 3). As a result of the very low noise level (~ pN), detailed observation of the mechanics of tube pulling is possible. A long pulling distance can be mandatory: T cells are rather small (~ 10 µm in diameter) but tethers can be pulled that are up to 10 or even 20 µm long, depending on the experimental condition.
Conclusions and perspectives
T cells have mechanosensory properties that range from their characteristic TCR molecule to the whole cell scale.2 We have presented here several examples which show how AFM, in imaging or force mode, is an highly interesting tool when used to characterize the surface which the T cells come in contact with, or the mechanics and forces of the T cells themselves.
AFM can also be combined with fluorescence microscopy to characterize, in real-time, some of the intracellular signals that are generated when the proteins present on the cell surface are stimulated. When used in combination with OT, a detailed characterization of cellular mechanics (Young's modulus, tension, and by using oscillating mechanical modes G' and G"-dynamic shear modulus) can be obtained. By varying the shape and size of the indentor over different scales, information on the sub-membrane organization of the cytoskeleton can also be obtained.
Very interesting possibilities arise when the systems are combined:
- Fluorescence and AFM or OT in real-time could for example lead to the characterization of the transfer function of the membrane/surface protein system.6 When AFM imaging is used in combination with fluorescence microscopy, 3D functional structures on the cell surface can be revealed.14
- Combining AFM with OT in a single system allows the reproduction of environments more similar to physiological ones, as one can assess the interaction between a T cell and an antigen presenting cell using SCFS, and when a helper, secondary T cell is brought into contact, the modulation of the recognition forces can be recorded.17 All in all, the use of nano-force tools is full of promise for the emerging fields of immuno-biophysics and immuno-mechanics. All in all, the use of nano-force tools is full of promise for the emerging fields of immuno-biophysics and immuno-mechanics.
Acronyms
APC: Antigen Presenting Cell
AFM: Atomic Force Microscope/y
(a)CD3: (antibody directed toward) Cluster of Differentiation 3, protein complex associated with TCR
(p)MHC: (peptide-loaded) Major Histocompatibility Complex
OT: Optical Tweezers
RICM: Reflection Interference Contrast Microscopy
SCFS: Single Cell Force Spectroscopy
SMFS: Single Molecule Force Spectroscopy
TCR: T Cell Receptor
TIRF(M): Total Internal Reflection Fluorescence (Microscope/y)
Authors
Anais Sadoun1 , Farah Mustapha1 , Omar Ndao1 , Gautier Eich1 , Yannick Hamon2, Laurent Limozin1, 3 , Pierre-Henri Puech1, 3
- Aix Marseille University, LAI UM 61, lnserm, UMR_S 1067, CNRS, UMR 7333, Turing Center for Living Systems, Marseille, F-13288, France.
- Aix Marseille Univ, CNRS, INSEAM, CIML, Marseille, France
- To whom correspondence may be sent: laurent.limozin@inserm.fr; pierre-henri.puech@inserm.fr
Aknowledgements:
This work was supported by funding from Prise de Risques CNRS (PHP), the ANR JCJC «DissecTion» (PHP), and from the Labex INFORM (ANR-11-LABX-0054, to lnserm U1067 Lab & CIML, and as PhD grants to AS & FM). This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sktodowska-Curie grant agreement No713750. Also, it has been carried out with the financial support of the Regional Council of Provence- Alpes-Côte d'Azur and with the financial support of the A*MIDEX (n° ANR- 11-IDEX-0001-02), funded by the lnvestissements d'Avenir project funded by the French Government, managed by the French National Research Agency (ANR) (as PhD grant to FM). It was also supported by GDR lmabio as grants to ON & GE.
References
- Murphy, Ken, Paul Travers, and Mark Walport. 2008. Janeway's lmmunobiology 7th Edition. Edited by Garland Science.
- Limozin, Laurent, and Pierre-Henri Puech. 2019. 'Membrane Organization and Physical Regulation of Lymphocyte Antigen Receptors: A Biophysicist's Perspective'. The Journal of Membrane Biology 252 (4-5): 397-472. https://doi.org/10.1007/s00232-019-00085-2.
- Wahl, Astrid, Celine Dinet, Pierre Dillard, Aya Nassereddine, Pierre-Henri Puech, Laurent Limozin, and Kheya Sengupta. 2019. 'Biphasic Mechanosensitivity of T Cell Receptor-Mediated Spreading of Lymphocytes'. Proceedings of the National Academy of Sciences of the United States of America 776 (13): 5908-73. https://doi.org/10.1073/pnas.1811516116.
- Puech, Pierre-Henri, Damien Nevoltris, Philippe Robert, Laurent Limozin, Claude Boyer, and Pierre Bongrand. 2011. 'Force Measurements of TCR/PMHC Recognition at T Cell Surface'. Edited by Daniel J. Muller. PLoS ONE 6 (7): e22344. https://doi.org/10.1371/journal.pone.0022344.
- Huse, Morgan. 2017. 'Mechanical Forces in the Immune System'. Nature Reviews Immunology 17 (11): 679-90. https://doi.org/10.1038/nri.2017.74.
- Cazaux, Severine, Ana'is Sadoun, Martine Biarnes-Pelicot, Manuel Martinez, Sameh Obeid, Pierre Bongrand, Laurent Limozin, and Pierre-Henri Puech. 2016. 'Synchronizing Atomic Force Microscopy Force Mode and Fluorescence Microscopy in Real Time for Immune Cell Stimulation and Activation Studies'. Ultramicroscopy 760 (January): 168-81. https://doi.org/10.1016/j.ultramic.2015.10.014.
- Lo Schiavo, Valentina, Philippe Robert, Zohar Mishal, Pierre-Henri Puech, Francesco Gentile, Paolo Decuzzi, Pierre Bongrand, and Laurent Limozin. 2013. 'Transient Adhesion Mediated by Ligand-Receptor Interaction on Surfaces of Variable Nanotopography'. International Journal of Nanotechnology 70 (5-7): 404-18. https://doi.org/10.1504/IJNT.2013.053512.
- Pi, Fuwei, Pierre Dillard, Laurent Limozin, Anne Charrier, and Kheya Sengupta. 2013. 'Nanometric Protein-Patch Arrays on Glass and Polydimethylsiloxane for Cell Adhesion Studies.' Nano Letters, June. https://doi.org/10.1021/nl401696m.
- Salles A. Billaudeau C, Serge A. Bernard A-M, Phelipot M-C, Bertaux N, et al. 2013. 'Barcoding T Cell Calcium Response Diversity with Methods for Automated and Accurate Analysis of Cell Signals (MAAACS)'. PLoS Comput Biol 9(9): e1003245. https://doi.org/10.1371/journal.pcbi.1003245.
- Dillard, Pierre, Rajat Varma, Kheya Sengupta, and Laurent Limozin. 2014. 'LigandMediated Friction Determines Morphodynamics of Spreading T Cells'. Biophysical Journal 107 (11): 2629-38. https://doi.org/10.1016/j.bpj.2014.10.044.
- Dejardin, Marie-Julie, Arnaud Hemmerle, Ana"fs Sadoun, Yannick Hamon, Pierre-Henri Puech, Kheya Sengupta, and Laurent Limozin. 2018. 'Lamellipod Reconstruction by Three-Dimensional Reflection Interference Contrast Nanoscopy (3D-RICN)'. Nano Letters, September. https://doi.org/10.1021/acs.nanolett.8b03134.
- Bufi, Nathalie, Michael Saitakis, Stephanie Dogniaux, Oscar Buschinger, Armelle Bohineust, Alain Richert, Mathieu Maurin, Claire Hivroz, and Atef Asnacios. 2015. 'Human Primary Immune Cells Exhibit Distinct Mechanical Properties That Are Modified by Inflammation'. Biophysical Journal 108 (9): 2181-90. https://doi.org/10.1016/j.bpj.2015.03.047.
- Hosseini, Babak H., Ilia Louban, Dominik Djandji, Guido H. Wabnitz, Janosch Deeg, Nadja Bulbuc, Yvonne Samstag, Matthias Gunzer, Joachim P. Spatz, and Gunter J. Hammerling. 2009. 'Immune Synapse Formation Determines Interaction Forces between T Cells and Antigen-Presenting Cells Measured by Atomic Force Microscopy'. Proceedings of the National Academy of Sciences of the United States of America 106 (42): 17852-51 https://doi.org/10.1073/pnas.0905384106.
- Puech, Pierre-Henri, Kate Poole, Detlef Knebel, and Daniel J Muller. 2006. 'A New Technical Approach to Quantify Cell-Cell Adhesion Forces by AFM.' Ultramicroscopy 106 (8-9): 637-644. https://doi.org/10.1016/j.ultramic.2005.08.003.
- Diz-Mufioz, Alba, Michael Krieg, Martin Bergert, ltziar lbarlucea-Benitez, Daniel J. Muller, Ewa Paluch, and Carl-Philipp Heisenberg. 2010. 'Control of Directed Cell Migration In Vivo by Membrane-to-Cortex Attachment'. Edited by William A. Harris. PLoS Biology 8 (11): e1000544. https://doi.org/10.1371/journal.pbio.1000544.
- Sadoun, Ana"fs, and Pierre-Henri Puech. 2017. 'Quantifying CD95/CI-CD95L Implications in Cell Mechanics and Membrane Tension by Atomic Force Microscopy Based Force Measurements'. Methods in Molecular Biology (Clifton, N.J.) 1557: 139-51. https://doi.org/10.1007/978-1-4939-6780-3_14.
- Chen, Jiahuan, Anutosh Ganguly, Ashley D Mucsi, Junchen Meng, Jiacong Yan, Pascal Detampel, Fay Munro, et al. 2017. 'Strong Adhesion by Regulatory T Cells Induces Dendritic Cell Cytoskeletal Polarization and Contact-Dependent Lethargy'. Journal of Experimental Medicine, 1-12. https://doi.org/10.1084/jem.20160620.