Imaging Neural Populations in Drosophila to Understand Natural Visual Processing
Discover innovative uses of two-photon imaging in visual neuroscience.
During this webinar, guest speaker, Max Turner, Ph.D. (Stanford University), discusses his latest work using two-photon microscopy to investigate the neural circuits that underlie computation and coding in the fly visual system. Gain first-hand insights into how visual neurons represent visual scenes and how multiphoton imaging opens up new possibilities for understanding visual processing.
Presenter's Abstract
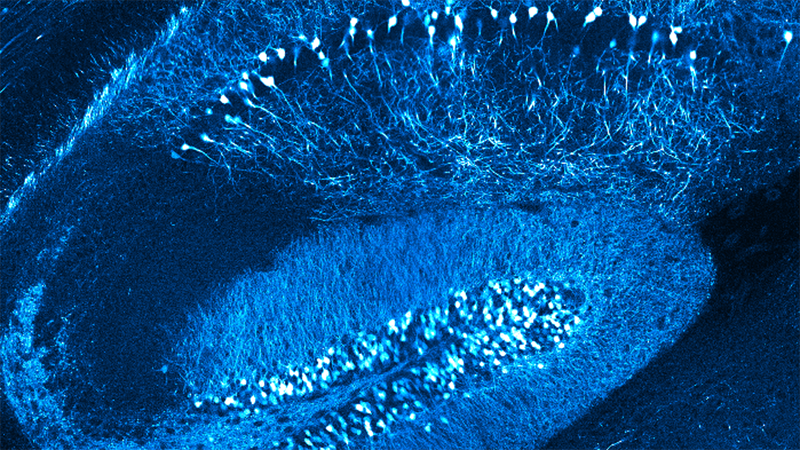
Natural vision is dynamic: as an animal moves, its visual input changes dramatically. How can the visual system reliably extract local features from an input dominated by self-generated signals? In Drosophila, diverse local visual features are represented by a group of projection neurons with distinct tuning properties.
Here we describe a connectome-based volumetric imaging strategy to measure visually evoked neural activity across this population. We show that local visual features are jointly represented across the population, and that a shared gain factor improves trial-to-trial coding fidelity. A subset of these neurons, tuned to small objects, is modulated by two independent signals associated with self-movement, a motor-related signal and a visual motion signal associated with rotation of the animal. These two inputs adjust the sensitivity of these feature detectors across the locomotor cycle, selectively reducing their gain during saccades and restoring it during intersaccadic intervals.
This work reveals a strategy for reliable feature detection during locomotion.
Find out more about the technology featured in this webinar or our other solutions for Imaging Neural Populations:
Speaker's
Max Turner, Ph.D., Postdoctoral Scholar at Stanford University
Max is a neurobiologist with an interest in the neural circuits that underlie visual computation and coding. He uses the Drosophila visual system as a model to study how diverse populations of visual neurons represent rich visual scenes, including those encountered during natural behavior.
He is currently a postdoctoral fellow in Tom Clandinin’s lab at Stanford University. He did his dissertation work in Fred Rieke’s lab at the University of Washington, studying how retinal ganglion cells in the mammalian retina encode natural scenes.
Kevin Mann, Ph.D., Applications Scientist, Bruker
Dr. Kevin Mann received Ph.D. degree from the University of California, Berkeley under the guidance of Dr. Kristin Scott where he studied fundamental behaviors in Drosophila using genetics, multiphoton microscopy, and electrophysiology. Next, he moved on to postdoctoral training in the laboratory of Dr. Tom Clandinin at Stanford University. Collaboratively he developed a method for whole-brain calcium imaging to detail the intrinsic functional neuronal network in Drosophila.