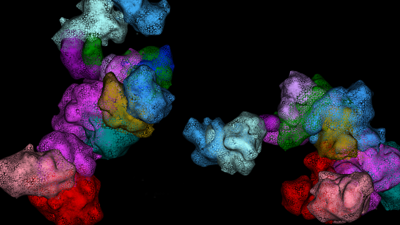
Notch-Mutated Cancer Genomes in 3D
The recording of this live event is unavailable
Unraveling Epigenetic Changes in Cancer Cells
During this live event, guest speaker Dr. Babak Faryabi, University of Pennsylvania, discussed utilizing Oligopaint and super-resolution microscopy to investigate the role of transcription hubs in altering genome folding. Attendees learned about the power of super-resolution microscopy and chromatin tracing in understanding the three-dimensional structure of the genome in single cells.
Presenter’s Abstract
The importance of epigenetic changes in cancer pathogenesis has become increasingly clear, yet the underlying mechanisms remain obscure. Our research focuses on identifying epigenetic changes that can render cancer cells addicted to specific transcriptional regulators. I will present evidence that oncogenic subversion of transcription factors leads to disruption of chromosome topology and instruct enhancer elements to aberrantly interact and activate intact key proto-oncogenes. These aberrant epigenetic states can drive cancer cell growth and proliferation in the absence of genetic mutations and may also confer drug resistance. I will also discuss our efforts to develop single-cell molecular tools and experimental models to characterize epigenetic mechanisms of oncogenesis and attendant therapeutic opportunities.
Find out more about the technology featured in this webinar or our other solutions for super-resolution microscopy:
Featured Products and Technology
Guest Speaker
Babak Faryabi, Ph.D., Associate Professor of Pathology and Cancer Biology, University of Pennsylvania Perelman School of Medicine
Dr. Faryabi has a distinguished history of research and accomplishments in cancer epigenomics. His work has significantly advanced our understanding of how transcription factors alter cancer genome folding. By conducting the first genome-wide and single-cell resolution measurement of genome looping in triple-negative breast cancer (TNBC) and mantle cell lymphoma (MCL), he discovered that Notch can regulate gene expression by changing genome folding and bringing together distant enhancer and promoter elements on the linear genome. This identified a novel mechanism by which oncogenic Notch alters transcription. Furthermore, he uncovered that Notch-dependent DNA loops bring multiple genomic elements into highly interacting spatial hubs of enhancers and promoters. He is among the first to elucidate the contribution of genome topology organization to cancer therapy resistance. His work revealed the role of genome folding reorganization as a novel epigenetic adaptation mechanism, conferring resistance to Notch inhibitors in T-cell leukemia and to BTK inhibitors in B cell lymphoma. To achieve his central scientific objective, he also innovates machine learning tools and software to better dissect the complexity and heterogeneity of genome structures and regulation in cancer.
As the faculty lead at the Center for Personalized Diagnostics at the Hospital of the University of Pennsylvania, his clinical expertise lies in developing genomic assays for diagnostic and predictive markers in cancer.