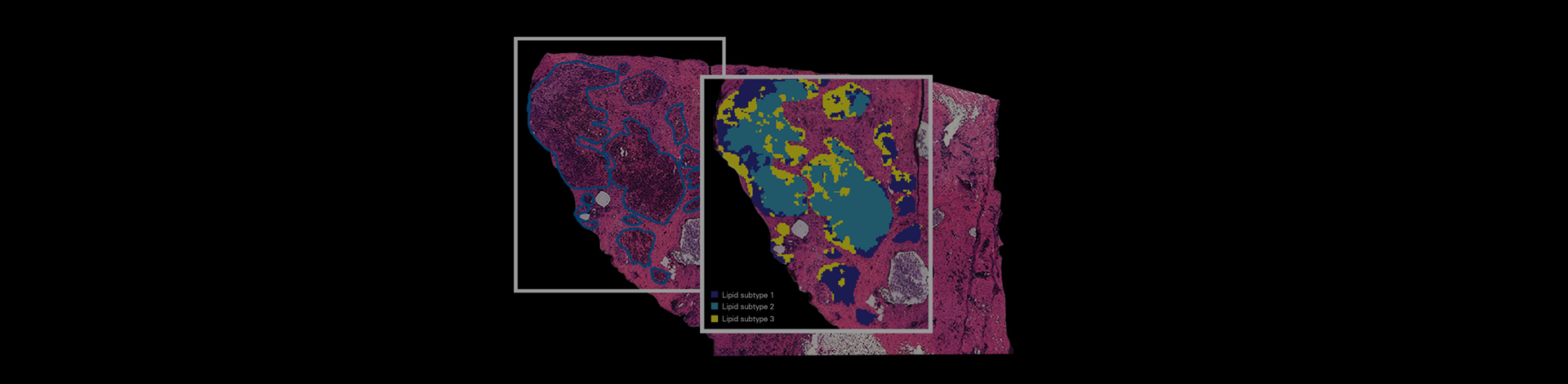

SpatialOMx® References
| Publication | Key Features | Research Focus | DOI |
|---|---|---|---|
| MALDI-2 on a Trapped Ion Mobility Quadrupole Time-of-Flight Instrument for Rapid Mass Spectrometry Imaging and Ion Mobility Separation of Complex Lipid Profiles. J. Soltwisch, et al., Anal.Chem (2020). | Lipid biology: Distribution and identification of lipid species. Bruker product: Bruker timsTOF fleX with MALDI-2 with SCiLS Lab MVS. Sample: Rat brain and testis. | Proof of concept study to show various lipid classes at high lateral resolution (10µm2) using MALDI-2 and TIMS. The combination of MALDI2 and TIMS successfully separated a variety of isomeric and isobaric ion species, helping to reveal further complexities of the lipodome. | DOI: /10.1021/acs.analchem.0c01747 |
| Mass‐Spectrometric Imaging of Electrode Surfaces—a View on Electrochemical Side Reactions. J. Fangmeyer, et al., Angew.Chem (2020). | Industrial applications: Identification and localization of compounds formed after electrode fouling. Bruker product: Bruker timsTOF fleX. Sample: Organic compounds on electrode surfaces. | Modern batteries undergo electrochemical side reactions, or “electrode fouling”. Polymerization of electrode surface compounds reduces efficiency. MALDI imaging is used for the first time to successfully visualize electrochemical side reactions. This technique can be applied as a tool for optimizing electro-organic reactions to benefit battery research. | DOI: /10.1002/ange.202010134 |
| Resolving the Complexity of Spatial Lipidomics Using MALDI TIMS Imaging Mass Spectrometry. K.V. Djambazova, et al., Anal.Chem (2020). | Lipid biology: Distribution and identification of lipid species. Bruker product: Bruker timsTOF fleX. Sample: Whole-body mouse pup. | Proof of concept study to enhance lipid isomer analysis by taking advantage of high mobility resolving power (>250). MALDI and TIMS combination allowed in situ separation of lipid isomers, with distinct spatial distributions, in whole mouse pups. | DOI: /10.1021/acs.analchem.0c02520 |
| Spatial Metabolomics of the Human Kidney using MALDI Trapped Ion Mobility Imaging Mass Spectrometry. E.K. Neumann, et al., Anal.Chem (2020). | Metabolite analysis: Identification and spatial distribution of small molecules. Bruker product: Bruker timsTOF flex. Sample: Human kidney tissue sections. | Low molecular weight compound analysis can be complicated by MALDI matrix ion interference. TIMS separation helps to resolve this. MALDI TIMS analysis at 20µm allowed detection of various metabolite classes from nutrients, biproducts, to food additives in the human kidney. | DOI: /10.1021/acs.analchem.0c02051 |
| In situ isobaric lipid mapping by MALDI–ion mobility separation–mass spectrometry imaging. T. Fu, et al., J.Mass Spectrom (2020). | Lipid Biology: in situ mapping and identification of lipids. Bruker product: timsTOF fleX and SCiLS Lab 2020a. Sample: freshwater crustacean Gammarus fossarum. | Proof of principle study to demonstrate high resolution imaging at 20µm with TIMS activated using a model organism. Distinct distributions of isobaric lipid species were observed in situ, demonstrating the specificity of this technique. | DOI: /10.1002/jms.4531 |
| Morphometric Cell Classification for Single‐Cell MALDI‐Mass Spectrometry Imaging. K. Ščupáková, et.al., Angew.Chem (2020). | Cell classification: Automated cell classification by combining morphometry and MALDI-imaging. Bruker product: timsTOF flex and rapifleX. Sample: Porcine colon and human gastric carcinoma tissue sections. | Morphometry is an enhanced histology technique that utilizes machine learning algorithms to automatically classify cells based on their morphology. MALDI imaging was done at 10 µm spatial resolution to acquire mass profiles of each cell type. These data were combined with morphometry to create matched mass profiles to classified cells. | DOI: 10.1002/anie.202007315 |
| MS Imaging‐Guided Microproteomics for Spatial Omics on a Single Instrument. F. Dewez, et. al., Proteomics (2020). | Spatial omics: Spatially guided proteomics based on lipid mass profiles on tissue. Bruker product: timsTOF flex and SCiLS Lab 2020a. Sample: Human breast cancer tissue sections. | MALDI imaging can provide spatially resolved mass spectra data on tissue but lacks the analytical capability to interrogate deep proteome. To obtain deeper omics data, MALDI-imaging was used to identify regions of interests in tissue samples. These regions of interest can be accurately removed by laser micro-dissection and analyzed by sensitive LCMS techniques. | DOI: 10.1002/pmic.201900369 |
| Rapid N-Glycan Profiling of Serum and Plasma by a Novel Slide Based Imaging Mass Spectrometry Workflow. C.R.K. Blaschke, et. al., J Am. Soc. Mass Spectr. (2020). | Glycan analysis: Identifying glycans from serum by MALDI-imaging and MS/MS. Bruker product: timsTOF flex, solariX MRMS and SCiLS Lab 2017a. Sample: Human serum. | N-glycans in serum and plasma has potential to serve as biomarkers for various diseases. Detecting and identifying glycans in serum spotted on slides by MALDI imaging and MS/MS was done as a proof of concept for high throughput analysis. This method was used for glycan profiling of breast cancer and benign lesion. | DOI: 10.1021/jasms.0c00213 |
| Detection of small molecule concentration gradients in ocular tissues and humours. B. Boughton, et al., J Mass Spectrom (2020). | Metabolite analysis: Spatial distribution of metabolites and small molecules. Bruker product: Bruker Solarix‐XR 7 T FT‐ICR mass Spectrometer & timsTOF fleX with SCiLS Lab Pro 2019. Sample: ocular tissue of various species (mouse, rat, rabbit, fish). | The spatial relationship of fluids and tissues in the eye allow it to effectively function, yet understanding of metabolic communication between these tissues is lacking. Tissue and ocular humour imaging performed in negative ion mode revealed common metabolite distributions across eye structures. A three-dimensional mouse eye analysis was also shown as proof-of-principle. | DOI: 10.1002/jms.4460 |
| Publication | Key Features | Research Focus |
| MALDI-2 on a Trapped Ion Mobility Quadrupole Time-of-Flight Instrument for Rapid Mass Spectrometry Imaging and Ion Mobility Separation of Complex Lipid Profiles. J. Soltwisch, et al., Anal.Chem (2020). DOI: /10.1021/acs.analchem.0c01747 | Lipid biology: Distribution and identification of lipid species. Bruker product: Bruker timsTOF fleX with MALDI-2 with SCiLS Lab MVS. Sample: Rat brain and testis. | Proof of concept study to show various lipid classes at high lateral resolution (10µm2) using MALDI-2 and TIMS. The combination of MALDI2 and TIMS successfully separated a variety of isomeric and isobaric ion species, helping to reveal further complexities of the lipodome. |
| Mass‐Spectrometric Imaging of Electrode Surfaces—a View on Electrochemical Side Reactions. J. Fangmeyer, et al., Angew.Chem (2020). DOI: /10.1002/ange.202010134 | Industrial applications: Identification and localization of compounds formed after electrode fouling. Bruker product: Bruker timsTOF fleX Sample: Organic compounds on electrode surfaces. | Modern batteries undergo electrochemical side reactions, or “electrode fouling”. Polymerization of electrode surface compounds reduces efficiency. MALDI imaging is used for the first time to successfully visualize electrochemical side reactions. This technique can be applied as a tool for optimizing electro-organic reactions to benefit battery research. |
| Resolving the Complexity of Spatial Lipidomics Using MALDI TIMS Imaging Mass Spectrometry. K.V. Djambazova, et al., Anal.Chem (2020). DOI: /10.1021/acs.analchem.0c02520 | Lipid biology: Distribution and identification of lipid species. Bruker product: Bruker timsTOF fleX Sample: Whole-body mouse pup. | Proof of concept study to enhance lipid isomer analysis by taking advantage of high mobility resolving power (>250). MALDI and TIMS combination allowed in situ separation of lipid isomers, with distinct spatial distributions, in whole mouse pups. |
| Spatial Metabolomics of the Human Kidney using MALDI Trapped Ion Mobility Imaging Mass Spectrometry. E.K. Neumann, et al., Anal.Chem (2020). DOI: /10.1021/acs.analchem.0c02051 | Metabolite analysis: Identification and spatial distribution of small molecules. Bruker product: Bruker timsTOF flex. Sample: Human kidney tissue sections. | Low molecular weight compound analysis can be complicated by MALDI matrix ion interference. TIMS separation helps to resolve this. MALDI TIMS analysis at 20µm allowed detection of various metabolite classes from nutrients, biproducts, to food additives in the human kidney. |
| In situ isobaric lipid mapping by MALDI–ion mobility separation–mass spectrometry imaging. T. Fu, et al., J.Mass Spectrom (2020). DOI: /10.1002/jms.4531 | Lipid Biology: in situ mapping and identification of lipids. Bruker product: timsTOF fleX and SCiLS Lab 2020a. Sample: freshwater crustacean Gammarus fossarum. | Proof of principle study to demonstrate high resolution imaging at 20µm with TIMS activated using a model organism. Distinct distributions of isobaric lipid species were observed in situ, demonstrating the specificity of this technique. |
| Morphometric Cell Classification for Single‐Cell MALDI‐Mass Spectrometry Imaging K. Ščupáková, et.al., Angew.Chem (2020). DOI: 10.1002/anie.202007315 | Cell classification: Automated cell classification by combining morphometry and MALDI-imaging Bruker product: timsTOF flex and rapifleX Sample: Porcine colon and human gastric carcinoma tissue sections | Morphometry is an enhanced histology technique that utilizes machine learning algorithms to automatically classify cells based on their morphology. MALDI imaging was done at 10 µm spatial resolution to acquire mass profiles of each cell type. These data were combined with morphometry to create matched mass profiles to classified cells. |
| MS Imaging‐Guided Microproteomics for Spatial Omics on a Single Instrument F. Dewez, et. al., Proteomics (2020). DOI: 10.1002/pmic.201900369 | Spatial omics: Spatially guided proteomics based on lipid mass profiles on tissue Bruker product: timsTOF flex and SCiLS Lab 2020a. Sample: Human breast cancer tissue sections | MALDI imaging can provide spatially resolved mass spectra data on tissue but lacks the analytical capability to interrogate deep proteome. To obtain deeper omics data, MALDI-imaging was used to identify regions of interests in tissue samples. These regions of interest can be accurately removed by laser micro-dissection and analyzed by sensitive LCMS techniques. |
| Rapid N-Glycan Profiling of Serum and Plasma by a Novel Slide Based Imaging Mass Spectrometry Workflow C.R.K. Blaschke, et. al., J Am. Soc. Mass Spectr. (2020). DOI: 10.1021/jasms.0c00213 | Glycan analysis: Identifying glycans from serum by MALDI-imaging and MS/MS Bruker product: timsTOF flex, solariX MRMS and SCiLS Lab 2017a. Sample: Human serum | N-glycans in serum and plasma has potential to serve as biomarkers for various diseases. Detecting and identifying glycans in serum spotted on slides by MALDI imaging and MS/MS was done as a proof of concept for high throughput analysis. This method was used for glycan profiling of breast cancer and benign lesion. |
| Detection of small molecule concentration gradients in ocular tissues and humours B. Boughton, et al., J Mass Spectrom (2020). DOI: 10.1002/jms.4460 | Metabolite analysis: Spatial distribution of metabolites and small molecules Bruker product: Bruker Solarix‐XR 7 T FT‐ICR mass Spectrometer & timsTOF fleX with SCiLS Lab Pro 2019 Sample: ocular tissue of various species (mouse, rat, rabbit, fish) | The spatial relationship of fluids and tissues in the eye allow it to effectively function, yet understanding of metabolic communication between these tissues is lacking. Tissue and ocular humour imaging performed in negative ion mode revealed common metabolite distributions across eye structures. A three-dimensional mouse eye analysis was also shown as proof-of-principle. |
For Research Use Only. Not for use in clinical diagnostic procedures.