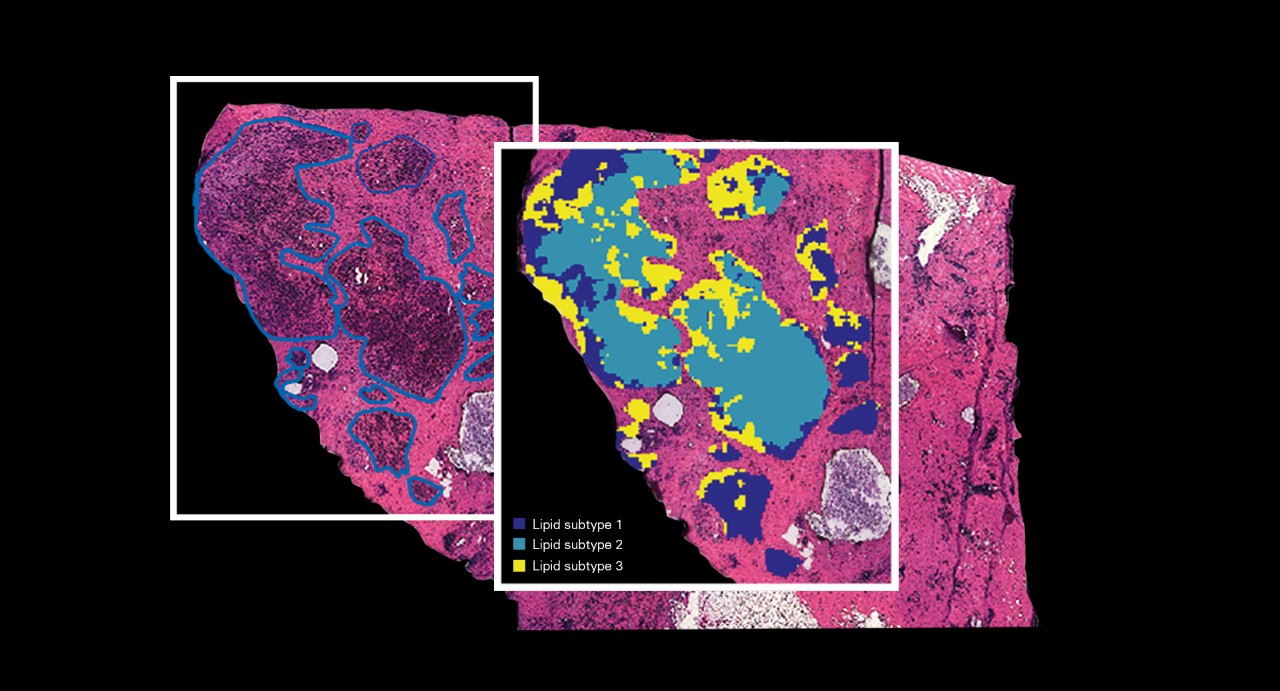

MALDI Guided SpatialOMx®
Reveal tumor cell subpopulations in greater molecular detail
Whether you are probing the cellular proteome, lipidome or metabolome, MALDI Imaging provides a literal map of molecular expression in the specimen that can be compared to reveal regional changes. Often, knowing where molecular expression changes can be just as important as knowing if expression changes. This can be especially true if certain compounds are highly spatially concentrated or if molecules co-distribute in specific compartments, vital information that is lost when examining only homogenized samples. MALDI Guided SpatialOMx® for untargeted discovery offers the most complete molecular range with contextualized spatial information providing important clues into intercellular communications networks that are integral to cancer growth.
Recent Studies
MS Imaging‐guided Microproteomics for Spatial Omics on a Single Instrument
Publication in Proteomics, August 7, 2020
Next level imaging Mass Spectrometry: Single cells in focus with SpatialOMx®
Prof. Ron Heeren, PhD, Maastricht University, Division Head Imaging MS, Maastricht, Netherlands
MALDI Guided SpatialOMx® uncovers proteomic profiles in tumor subpopulations of breast cancer
SpatialOMx® is providing context to OMICS - Learn more in this interview with Michael Easterling:
What’s one of the most important prerequisites for MALDI Imaging? LC-MS! Find out how SpatialOMx® is providing context to OMICS and where the field is going next and why pathology researchers care in an interview with Michael L. Easterling, Manager of Bruker’s Imaging Business.
Grant Support Materials
Brochure: timsTOF Pro/fleX The new standard for shotgun proteomics
Four reasons to switch to 4D-Proteomics™ on the timsTOF platform. To provide concise arguments for replacement of older 3D mass spectrometers with the 4D capable, timsTOF platform and its unique TIMS (Trapped Ion Mobility Spectrometry) capability which adds the CCS dimension to retention time, precursor mass, and MS/MS.
For more insight into MALDI Guided SpatialOMx®, listen to our experts in Bruker’s On-Demand webinars:
SpatialOMx® – bringing context to proteomics through molecular pathology
Dr. Shannon Cornett, MALDI Imaging Business Unit, Bruker Daltonics
Imaging pathways: SpatialOMx® captures small molecule and lipid networks across tissues to bridge metabolomics and disease
Dr. Shannon Cornett, MALDI Imaging Business Unit, Bruker Daltonics
In situ image identification using MALDI
Dr. Janina Oetjen, Application Specialist MALDI Imaging, Bruker Daltonics
For further information, please contact:
Sales:
ms.sales.bdal@bruker.com to arrange for a demo or to contact sales
For Research Use Only. Not for use in clinical diagnostic procedures.