
MBT and Mycobacteria Identification
MALDI Biotyper® navigating the mycobacteria identification challenge
Equip your MALDI Biotyper® with the MBT HT Mycobacteria Module to unveil a smooth workflow for mycobacteria identification. This package encompasses a specific reference spectrum library and a software module, empowering you to navigate the mycobacteria landscape with confidence.
spectra from mycobacteria
no boiling required for deactivation
Solve the mycobacteria identification puzzle
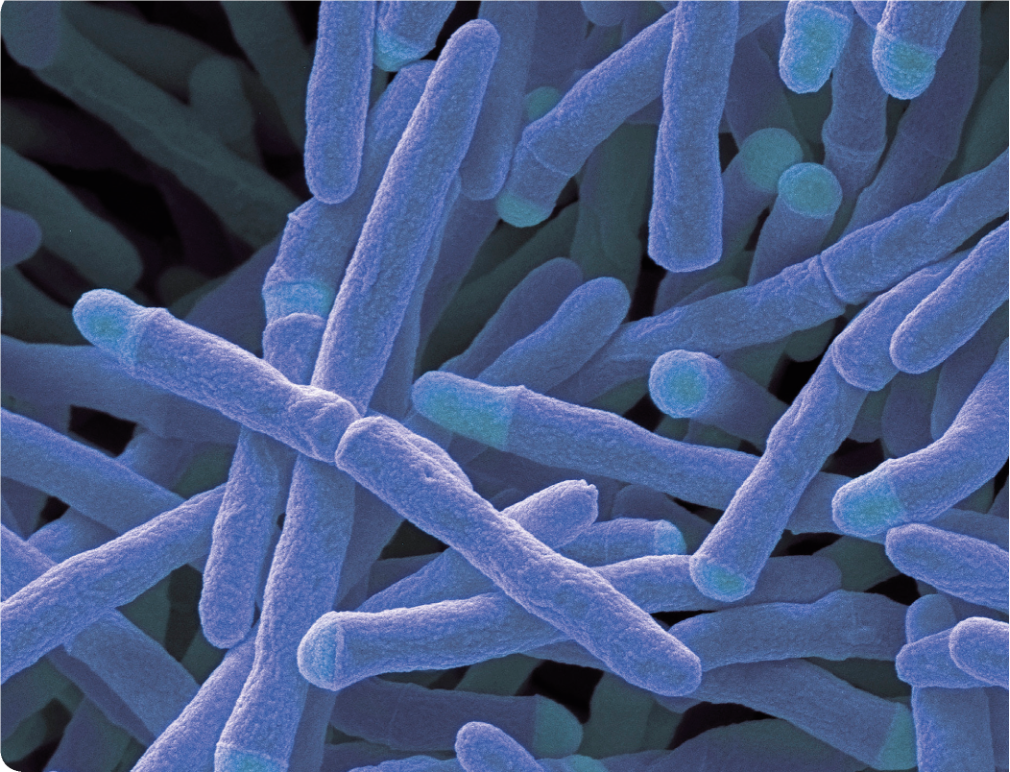
Within the genus Mycobacterium lies the intricate world of the Mycobacterium tuberculosis complex (MTC) and the nontuberculous mycobacteria (NTM), including opportunistic pathogens. As NTM-related infections surge and antibiotic resistance climbs, the demand for accurate differentiation among mycobacteria species has never been more critical.
Antimicrobial susceptibility differences underscore the urgency of species-level mycobacteria identification. Yet, traditional methods, often plagued by lengthy turnaround times and inaccuracies, have struggled to keep up.
While molecular methods like nucleic acid hybridization probes quickened the pace, they occasionally fell short in specificity. Sequencing target genes like 16S rDNA offered speed and accuracy but often at a steep price, limiting accessibility.
Rely on the MBT Mycobacteria Module
Expand your MALDI Biotyper’s potential to include mycobacteria identification with the MBT HT Mycobacteria Module, which holds a specific reference spectrum library and a software module.
The MBT Mycobacteria Library, housing reference spectra for 182 out of the current 201 known mycobacteria, ensures mycobacteria sample analysis regardless of growth on liquid or solid medium.
The dedicated software module processes the typically more complex mass spectra originating from mycobacteria, offering insights beyond conventional methods. Samples flagged as mycobacteria samples undergo an optimized acquisition process, automatically comparing spectra to the MBT Mycobacteria Library for precise, sensitive identification.
Introducing the MBT Mycobacteria Kit
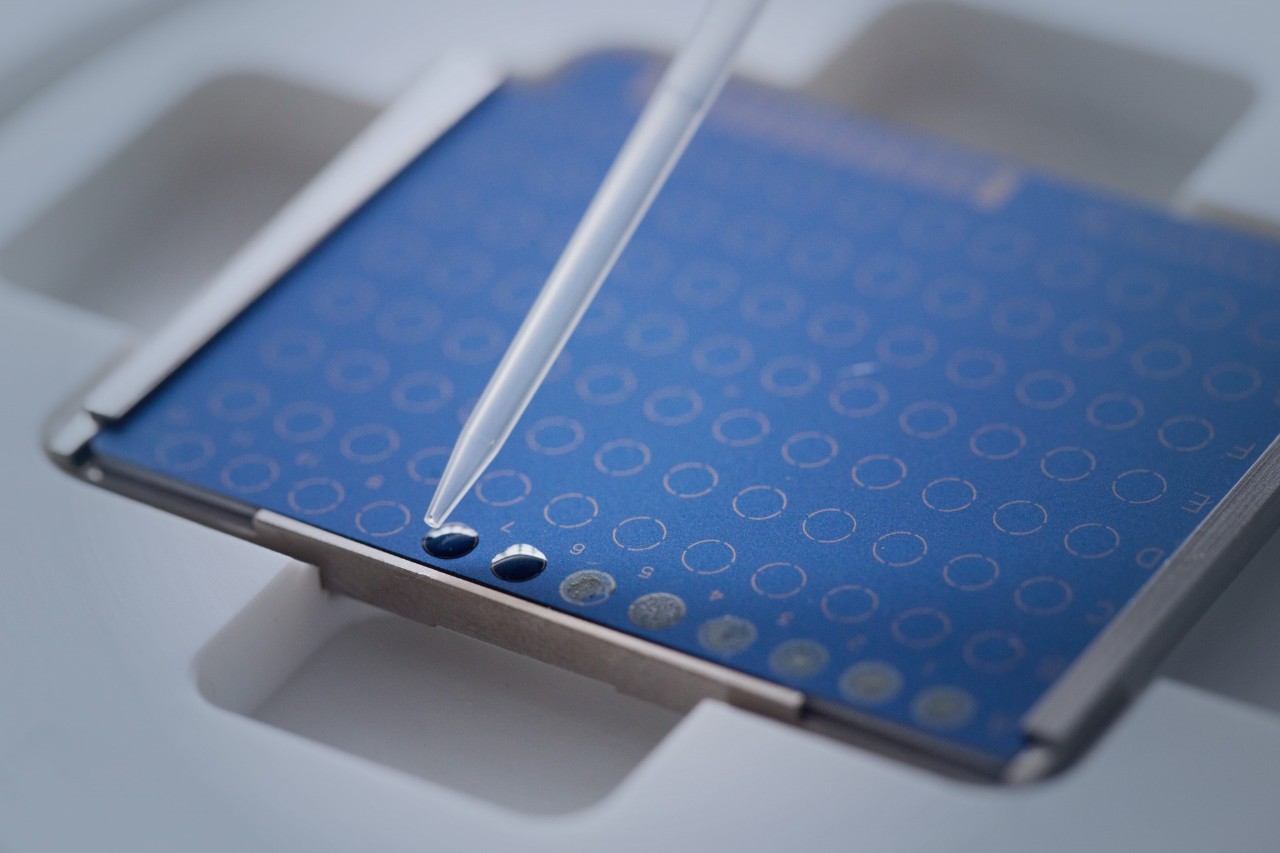
In the pursuit of excellence, we introduce the innovative MBT Mycobacteria Kit. Tackling the challenges posed by mycobacteria, this kit redefines their sample preparation. Unlike other methods, our unique inactivation process, involving an Inactivation Reagent to be used at room temperature, and mechanical disruption of cell aggregates, replaces cumbersome boiling. This advanced method ensures biosafety level 3 organisms are handled securely.
The MBT Mycobacteria Kit transforms sample preparation for Mycobacterium spp., whether cultivated in liquid or solid media.
Workflow of the MBT Mycobacteria Kit
Mycobacteria solutions fitting to your microbiology lab
The mycobacteria solution is available in two different versions. Please contact your local representative for availability in your country:
MBT HT Mycobacteria Module
The MBT HT Mycobacteria Module includes a software module and a dedicated mycobacteria library. The MBT Mycobacteria Kit ensures safe and fast sample preparation.
Not for use in clinical diagnostic procedures.
MBT HT Mycobacteria IVD Module
The MBT HT Mycobacteria IVD Module includes a software module and a dedicated mycobacteria library. The MBT Mycobacteria IVD Kit ensures safe and fast sample preparation.
For professional use only. Not for sale in the USA.