Toward Quantitative Nanomechanical Measurements on Live Cells with PeakForce QNM
Live cells mechanics measurement with PeakForce QNM
Measuring and mapping mechanical properties of live cells is of high importance in today’s biological research. Atomic force microscopy1 has been recognized since the mid-eighties as an excellent technique to image a wide range of samples in their near-natural environment. Although the primary function of atomic force microscopy is to generate a three-dimensional (3D) profile of the scanned surface, much more information is available through the technique. TappingMode™, which was developed in 1993,2 prevents tip and sample damage from friction and shear forces, and allows qualitative mechanical property mapping through phase imaging. Around the same time, force spectroscopy,3 and force volume4 were developed to study tip-sample forces at a point, or over an area respectively. Traditionally, force spectroscopy and force volume are the most commonly used modes to quantitatively measure mechanical forces at the nanometer scale. Unfortunately, both techniques have suffered from slow acquisition speed and a lack of automated tools to analyze the hundreds to thousands of curves required for good statistics.
The recent release of PeakForce QNM® resolves these limitations and can provide better results in terms of resolution, speed, ease-of-use, and quality of delivered information.5 PeakForce QNM is based on Bruker’s PeakForce Tapping™ mode, which oscillates the probe at about 1kHz, and which uses the peak force (maximum nominal force applied to the sample) for feedback control. Each time the tip interacts with the sample, a force curve is collected and analyzed by PeakForce QNM. The resulting signals are extracted and quantitatively displayed as a falsecolored image in real-time. Currently available data types include peak force, adhesion, Young’s modulus, deformation and dissipation. This mode has been successfully tested on a wide range of samples,6 from bio-polymers7,8 and live eukaryotic cells9 to human models.10
This application note reviews recent progress in mapping the properties of soft samples such as cells and gels with force volume and PeakForce QNM and the use of the newest NanoScope® and NanoScope Analysis features to collect and analyze the data from these techniques.
Force Volume and PeakForce QNM:
Speed and Flexibility
Force spectroscopy, force volume and PeakForce QNM are all useful methods for studying cell mechanics. Figure 1A summarizes the characteristics of each technique and compares their principle benefits and drawbacks. PeakForce QNM is best for high-resolution imaging or relatively high-speed imaging where material properties are of interest. Force volume is useful as a comparison to PeakForce QNM and for cases where the loading rate dependence of the measurement is important (such as studies of viscoelastic behavior and kinetic binding or unfolding measurements). Single force spectra are best for situations where mapping is not as important, where there are only a few points of interest in a sample, or where force measurements need to be separated by some distance to avoid interaction from one measurement to the next. The key differences between PeakForce QNM and force volume (or force spectroscopy) are:
- PeakForce QNM uses a sinusoidal modulation of the base of the cantilever relative to the sample surface, while force volume uses linear ramping. This allows PeakForce QNM to acquire thousands of ramps per second where force volume ramping is usually limited to a maximum of around ten ramps per second. The higher ramp rate of PeakForce QNM enables the acquisition of more detailed material property maps in much less time than force volume.
- PeakForce QNM controls the normal force of the tip-sample interaction by detecting the peak force of each tap and feeding the information into a feedback loop that is running continuously. The force control benefits from the results of previous taps and from the fact that the sinusoidal waveform causes the tip velocity to approach zero as the tip approaches the peak, allowing for ultra-low interaction force (as low as 10 piconewtons). In contrast, force volume treats each tap separately, and the tip approaches the surface at full speed. Once the trigger force is detected, the system attempts to retract instantaneously, leading to overshoot and larger forces, especially at higher ramp rates.
- PeakForce QNM is usually also more stable than force volume. This is because there is less time for the system to drift within an image and because the superior force control inhibits tip wear and contamination.
For these reasons, PeakForce QNM is significantly better for material property mapping than force volume in most cases. On the other hand, force volume and force spectroscopy allow more control of the details of the ramp, such as independent control of approach and retract tip velocity and surface hold. Also, force volume and force spectroscopy taps can be separated by some distance since the triggering is treated independently, while PeakForce QNM taps must be close together to achieve the best feedback performance.
Figure 1 also describes some of the new features of Bruker’s recently introduced Nanomechanics package. This package adds key functionality that allows users to easily bridge between the techniques of PeakForce QNM, force volume and single force spectra, while adding some important capabilities for working with soft materials such as cells.
For PeakForce QNM, the nanomechanics package adds the capability to work at a wider range of frequencies and amplitudes. This is useful for working in fluid (smaller frequencies and amplitudes reduce viscous drag on the cantilever) and for very soft samples (where larger amplitudes may be needed). In response to numerous requests from the research community, the Sneddon cone model of elastic deformation was also added.11 This allows calculation of modulus based on a conical or pyramidal tip shape instead of the parabolic (spherical) tips of the DMT model. On very soft samples (e.g., cells, tissues,biomolecules), the tip often indents well past the part that can be approximated by a sphere (see Figure 1B), even with the best force control. Thus, the Sneddon model is often more appropriate for biological samples. Finally, PeakForce Capture™ adds the capability to simultaneously save a force curve at every pixel in the image while collecting PeakForce QNM material property maps. The data is saved in a force volume type file, enabling easy comparison to force volume images with the same analysis functions. Data channels can be recalculated with different parameters (deflection sensitivity, etc.) or models, or individual curves can be examined and compared.
For force volume, it is now possible to obtain maps of DMT modulus, Sneddon modulus, adhesion, and peak force in real-time in addition to the traditional force slice. This provides immediate confirmation that the experiment is going well, or needs adjustment, without time consuming offline analysis. Once the data is collected, the offline analysis view allows the material properties to be recalculated with different parameters (deflection sensitivity, etc.) or models, or individual curves can be examined, compared, and exported to the single curve format.
Some new tools are also available for single force spectra (see Figure 1C). Modify force parameters allows the calibration of individual curves to be adjusted. Baseline correction and Boxcar filter allow the curves to be filtered to correct artifacts such as offset, tilt, and noise. Indentation analysis allows fitting of the curves with either DMT model or Sneddon with options to include adhesion, use approach or retract, etc. All of these functions can be automated to easily analyze hundreds or thousands of curves and generate reports with statistics or histograms of data. Finally, for more complex analysis, a new MATLAB toolbox allows MATLAB to directly access NanoScope data files, freeing researchers to focus on modeling and results instead of worrying about file parsing or ASCII exports.
The new nanomechanics package provides all of the tools necessary for easy comparison between the different techniques, to test different models, and to explore time dependence of the tip-sample interaction. This can be done either at a single point or in a map of the variation in material properties across a sample surface.
Preliminary Testing on Bacteria
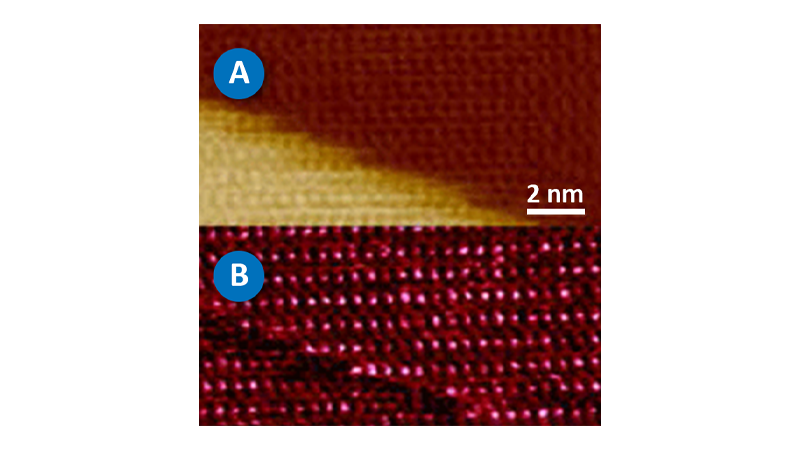
Figure 2A shows a pair of E. coli bacteria rendered in 3D (based on the topography of the sample) with the brightness of the image based on the Young’s modulus.
This image was collected using PeakForce QNM in about 10 minutes. This is in contrast to the force volume image in Figure 2C, which took about 35 minutes to collect. The improvement in speed is convenient and allows more and higher resolution images (256x256 vs. 64x64 in this case) to be collected during the life of a cell. The bacteria are still alive during imaging (in liquid at room temperature) and the one on the right appears to be in the process of dividing. Note that the dividing cell on the right is significantly softer than the cell on the left (~2MPa vs. ~15MPa). Much of the substrate is too stiff to accurately measure under the same conditions as the bacteria, but note the presence of some softer components, including the bacterial flagella in the lower-right corner. For these images, the modulus was calculated using the Sneddon (conical) model assuming a tip with an 18 degree half angle (typical for the DNP-A type of probe that was used).
It is often useful to examine individual force curves from interesting parts of the image. PeakForce Capture allows the simultaneous capture of the force curves that are used to create the property maps in PeakForce QNM. As before, the images are calculated and captured normally, but a force curve for each pixel in the property map is saved separately in a Quantitative Force-Volume file format. These files can be opened in NanoScope analysis for additional exploration with the full Quantitative Force-Volume analysis tools. It is easy to examine or export individual curves or sets of curves. Additionally, calibration parameters can be adjusted and different property models can be compared (e.g., DMT vs. Sneddon). Since the same tools are used to analyze both force volume and PeakForce Capture files, it is simple to contrast results collected at ramp rates from 0.1-10Hz (force volume) to those at ramp rates from 250Hz and higher (PeakForce Capture). This allows investigation of the time dependence of the tip sample interaction.
Figure 2B shows a pair of force curves (approach only) from PeakForce Capture. One was collected on top of the cell, while the other was collected on the sample substrate as indicated in Figure 2A. The slope of the curve from the substrate (red) is initially smaller than that of the bacteria, but it increases rapidly, becoming nearly vertical at the most extended point. This occurs because there is a soft, thin gelatin coating on the substrate that is intended to aid immobilization of the bacteria cells. When the tip penetrates deeper into the gelatin, it begins to feel the underlying glass substrate, which is much stiffer than the cells. The curve collected on the cell (green) is more uniform in slope, indicating that the substrate is not influencing the measurement significantly. The ability to access the individual curves creates significant opportunities for additional exploration and understanding.
Probing Agarose Gels at Various Ramp Rates
Agarose is a polysaccharide derived from agar. It is most widely used as a medium for gel electrophoresis measurements but it is seeing more recent applications as a tissue mimicking material (e.g., tissue phantoms).12–15 The mechanical properties of agarose gels are concentration dependent, with higher agarose concentrations resulting in “stiffer” gels. This allows preparation of agarose gels having different elastic modulus values in the biologically relevant range of tens to thousands of kilopascals. To demonstrate the ability of force volume and PeakForce QNM to measure these very soft samples, gels from 1%-5% wt. were prepared and imaged in PBS buffer. In Figure 3A, typical approach curves collected on the 3% agarose sample with ramp rates from 1Hz (force volume) to 250Hz (PeakForce QNM) are overlaid on the same plot to demonstrate the similarity between results from the two techniques, even over several orders of magnitude of ramp rate. If Sneddon modulus is calculated for each of these curves, the results are 1Hz: 353kPa, 5Hz: 340kPa, 10Hz: 357kPa, 250Hz: 351kPa. These results are essentially identical within the uncertainty of the measurement.
Figure 3C shows the same thing in a more statistically relevant way. The force volume results (1Hz) are shown in green with the PeakForce QNM results at 250Hz in red. The PeakForce QNM histogram provides much better statistics since it is based on 16,384 (128x128) measurements compared to 256 (16x16) measurements for force volume. Despite the large difference in number of samples, the acquisition time is similar: ~11 minutes for PeakForce QNM vs. 4 minutes for force volume. Thus, PeakForce QNM provides significantly better statistics than force volume, at minimal cost. The Young’s modulus can be calculated by directly analyzing the color contrast of the captured images (bearing analysis) or by exporting and processing the selected force curves, using the NanoScope Analysis Force package features (see Figure 1C). When thousands of curves are involved, automated analysis becomes critical.
If the sample has a significant time-dependent deformation mechanism, such as viscoelasticity, the results of force volume and PeakForce QNM may be different. This is because the time dependence allows different amounts of deformation at the time scale of the ramp. In Figure 3(B-D) we see that there is very little difference between measurements at 250Hz (PeakForce QNM) and 1Hz (force volume) for the 1% and 3% gels, but the 5% gel has some increase in modulus at higher frequencies. Note that the Sneddon and DMT models only include elastic deformation mechanisms (without time dependence). To quantify this time-dependent behavior, new analysis techniques are needed. The new MATLAB toolbox in NanoScope Analysis is a great tool for researchers interested in developing these techniques.
PeakForce QNM to Study Plant Morphogenesis
Morphogenesis in biology (i.e., development of an organism’s shape) is a complex mechanism controlled by biochemical factors, themselves dependent on gene expression, and on biophysical factors. Unlike animals where morphogenesis is based on cell division, cell growth, cell migration and apoptosis, plant morphogenesis in young tissues only results from cell division and elongation. The shape of plant cells is governed by the turgor pressure inside the cells and by the mechanical anisotropy and stiffness of the extra-cellular matrix (cell wall). Because of this relative simplicity, plants are ideal systems to study the role of nanomechanics in the morphogenesis of multicellular organisms.
In plants, morphogenesis mostly depends on a pool of stem cells localized in specialized plant tissues called meristems, which determine the shape and location of organs (flowers and leaves) and keep the plants growing. Meristems can produce organs over the entire lifetime of the plant, and are physically accessible. Because they control the number, type, and position of the lateral organs, meristems also largely determine key agronomic features such as fruit size, biomass, flowering period, and leaf number. Therefore, getting a better insight into their mechanical properties at the micro- or nanometer scale is of high interest, both for fundamental research and in the agribusiness.16–18
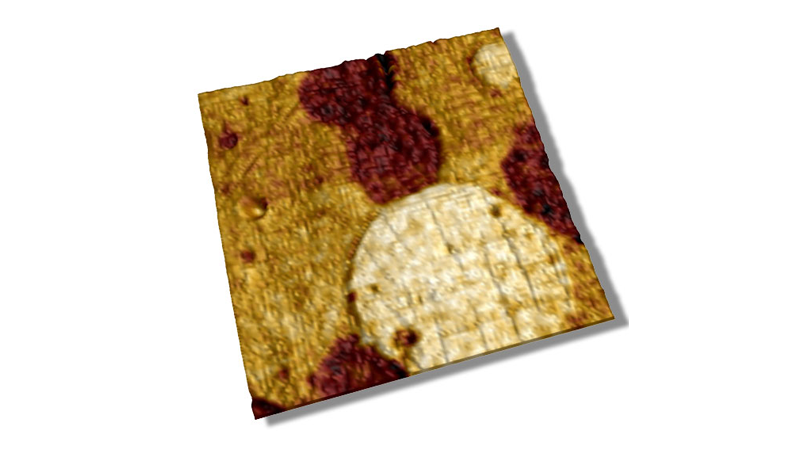
Meristem cells were imaged by confocal optical microscopy integrated with the BioScope Catalyst™, and an area of interest was selected to perform the AFM measurements (rectangular area in Figure 4A) through the MIRO® software interface. Anticlinal cell walls (normal to the surface) are thought to be much stiffer than the rest of the cells. Fluorescence labeling of the cell walls with a lipophilic dye allowed easy identification of their location within the meristem for correlation to AFM modulus measurements (see Figure 4A). The same meristem cells were imaged in PeakForce QNM mode. This type of tissue is quite bumpy, with many relatively tall features (up to 10μm) but it can easily be imaged using Bruker’s ScanAsyst® mode (also based on PeakForce Tapping technology) with full automatic optimization of imaging parameters. In Figure 4B, the topography in the area of interest is rendered in 3D showing ~50 cells. In Figure 4C the 3D topographic surface was recolored based on the elastic modulus map of the area (softer areas are darker), clearly indicating that the cell walls are much stiffer than the cell core. While moduli can be influenced by the topography of a surface (valleys sometimes appear to be stiffer than peaks because the contact area with the tip is greater), this was not a significant bias in this measurement. Indeed, green circles show areas where there are divided cells presenting newly synthesized cell walls observable in the confocal and modulus maps, but not in the topographic map. This type of study illustrates the potential of PeakForce QNM to investigate the mechanical changes of plant cell walls during development, opening possibilities to relate local biophysical parameters to the global shape of the tissue in the presence of hormones, drugs, or in a modified genetic background.19
Force Volume vs. PeakForce QNM on Live Cells
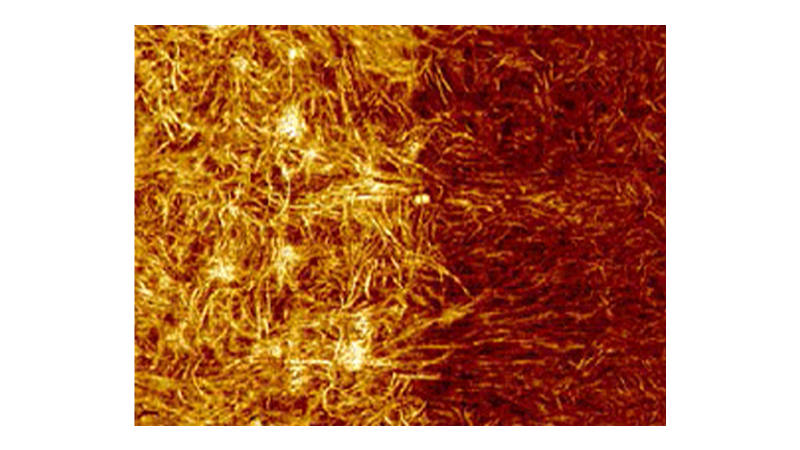
Many phenomena relating to the lifecycle and behavior of cells and tissues are related to cell mechanics or the mechanics of the constituent parts of the cell. Force volume and PeakForce QNM mapping are both valuable tools when investigating phenomena such as cell migration and cell division or when using cell mechanics to detect cancer or disease. The faster, higher resolution mapping made possible with PeakForce QNM allows more detailed mapping of these processes as they occur. The limited duration of these processes and cell lifetime make speed of acquisition a critical component of many experiments. Figure 5 demonstrates the resolution possible on the lamellipodium of a mouse B16 cell imaged in HEPES buffer. This image was collected in 8.5 minutes. A similar force volume map collected at a ramp rate of 2Hz with the same resolution would take about 9 hours.
Figure 5B and 5C compare modulus maps calculated from the DMT (sphere) model using the retract curves and the Sneddon (cone) model using the approach curves. The substrate (upper right of both images) is saturated since the cantilever spring constant of the probe used is too small to provide quantitative modulus on the much stiffer surface of the glass bottom Petri dish. The DMT modulus map is brighter than in the Sneddon map, with the Sneddon map being more accurate since the deformation depth on the cell was between 100 and 200nm (tip radius is ~30nm, so the tip shape can be approximated by that of a cone). To demonstrate the difference, a single curve was extracted from the PeakForce Capture data and analyzed with both the DMT model (B) and the Sneddon model (C). Comparing the green fit curve to that of the data, it is easy to see that the Sneddon model fits the data much better than the DMT model.
During image acquisition, force curves (similar to those in Figure 5B and 5C) were collected and analyzed along with the topography, providing the simultaneous maps of DMT modulus, Sneddon modulus, peak force error, deformation, dissipation, and adhesion. Figure 5D shows six more of these maps. Details such as the actin fibrils in the cell cytoskeleton are visible in many of the channels, but there are differences. For example, the peak force error channel and deformation channel show many small fibrils, while fewer fibrils are visible in the modulus channels, and none are visible in the adhesion map.
While PeakForce QNM can correct for some background deflection not related to the short range tip-sample interaction, it is not always possible to correct all of the background in fluid due to variation of apparent liquid viscosity near the sample surface. For highest measurement accuracy, it is best to minimize this viscous background by using the smallest modulation amplitude possible and <1kHz modulation frequency. These images were collected with modulation amplitude of 200nm at 250kHz. The probe was also selected with the viscous background in mind. The classic MLCT-D probes have fairly small cantilever surface area, with a moderately low spring constant of 0.048N/m. Tip geometry was pyramidal with 35 degree half angle and R~30nm end radius.
Force volume is well accepted as a tool for studying the mechanical properties of cells at ramp rates of less than 10–20Hz. Figure 6 compares the Sneddon modulus maps from Figure 5 with those from force volume at 1Hz and 5Hz, and with PeakForce Capture (PFC) results at 250Hz. All of the maps show that the cell’s lamellipodium has about the same modulus (compare the color of the lower left part of each image in 6A and 6C) independent of ramp rate and technique. Figure 6B shows a histogram of all of the images, showing that all of the peaks are around 20kPa. The lack of variation indicates that there is very little viscoelasticity or any other time-dependent deformation mechanism active in the range of frequency between 1Hz and 250Hz for the lamellipodium of this cell.
The force volume images were collected at low resolution (16x16 pixels) to save time, but the resulting modulus maps do not have enough resolution to clearly identify the actin fibrils in the cytoskeleton that are visible in the PeakForce Capture and PeakForce QNM images.
PeakForce QNM Investigation of Cancer Cells
It’s currently accepted that cancer cells are usually softer and more deformable than their normal counterparts.20–22 Glioblastoma is the most aggressive and common form of malignant brain cancer, and is one of the most difficult to treat forms of cancer because the tumor cells are resistant to available therapies and because few drugs can cross the blood-brain barrier to act on the tumor. U-251 is an isogenic cell line of glioblastoma that is extremely invasive.
This tendency to invade can be suppressed by overexpressing some tumor suppressive factors (TSFs). This modification is also expected to induce changes in mechanical properties. Those changes have been investigated in PeakForce QNM mode and the results are reported in Figure 7.
PeakForce QNM can either be used to image challenging cells at a high resolution (Figure 7A and 7B: 20x20μm images showing sharp details of the cell cytoskeleton) or to sense the mechanical properties (Figure 7C, 7D and 7E show peak force error, Young’s modulus and deformation in 80x80μm images of U-251 live cells over-expressing a TSF). Those images have a 128x128 resolution. Using Adaptive Scanning (a feed-forward control allowing dramatic improvement in XY positioning for large scan sizes), allows a capture time of 5 to 6 minutes. Force volume and PeakForce QNM have been tested on a high number of wildtype and TSF-transfected cells. The results can be summarized as follows:
- Both force volume and PeakForce QNM clearly demonstrate that the transfection causes the cells to become much stiffer (and less deformable), as expected.
- The standard deviations on the Young’s modulus for these measurements are significantly higher in force volume than in PeakForce QNM measurements. Additionally, there are significantly more data points for analysis with PeakForce QNM than with force volume. Together this allows for a much lower error standard error of the mean (assuming the variations in the measurement are statistically independent, SEM=σ/√n) and more accurate results. For an average image capture time of 5 minutes PeakForce QNM can capture nine 256x256-pixel images per sample type, providing 589,824 force curves, each analyzed to obtain a modulus data point. In contrast, force volume can only obtain 32x32-pixel images in the same amount of time, resulting in 9,216 data points.22 Even if the standard deviations of the two samples were the same, the PeakForce QNM case would have a standard error of the mean of about eight times lower than that for force volume.
For this type of measurement, PeakForce QNM is much more relevant than force volume in terms of resolution, quality, and amount of delivered information, and thus offers great perspectives in cell mechanics investigation.
Summary
The mechanical properties of biological samples often affect their structure and functional activity and are, hence, very important to biologists. Force volume has been accepted since the mid-1990s as a powerful tool for measuring and mapping the mechanical properties of biological samples. Force volume is optimized for mapping with low ramp rates (~0.5–10Hz) and relatively low resolution. PeakForce QNM improves upon force volume in terms of resolution and speed (with ramp rates ~250Hz–2KHz), making it more practical to collect and analyze much more data for better detail and statistics. Together, force volume and PeakForce QNM provide new opportunities for comparisons of material response at over about four orders of magnitude of ramp rate in air or liquid environments. In addition, the new features of NanoScope and NanoScope Analysis offer the user a maximum of ease-of-use and flexibility to collect, process, and analyze the thousands of force curves in a typical force volume or PeakForce QNM map.
References
- G. Binnig, C.F. Quate, and C.Gerber, “Atomic Force Microscope,” Phys. Rev. Lett. 56 (1986) 930–33.
- Q. Zhong, D. Innis, K. Kjoller, and V.B. Elings, “Fractured Polymer/Silica Fiber Surface Studied by Tapping Mode Atomic Force Microscopy,” Surf. Sci. 290 (1993) 688–92.
- G.U. Lee, D.A. Kidwell, and R.J.Colton, “Sensing Discrete Streptavidin Biotin Interactions with Atomic- Force Microscopy,” Langmuir 10 (1994) 354–57.
- M. Radmacher, J.P. Cleveland, M. Fritz, H.G. Hansma, and P.K. Hansma, “Mapping Interaction Forces with the Atomic Force Microscope,” Biophys. J. 66, (1994) 2159–65.
- B. Pittenger, N. Erina, and C. Su, “Quantitative Mechanical Properties Mapping at the Nanoscale with PeakForce QNM,” Bruker Application Note #128 (2011).
- A. Berquand, “Quantitative Imaging of Living Biological Samples by PeakForce QNM,” Bruker Application Note #135 (2012).
- J. Adamcik, A. Berquand, A., and R. Mezzenga, “Singlestep Direct Measurement of Amyloid Fibrils Stiffness by Peak Force Quantitative Nanomechanical Atomic Force Microscopy,” Appl. Phys. Lett. 98 (2011) 193701–03.
- T.J. Young, M.A. Monclus, T.L. Burnett, W.R. Broughton, S.L. Ogin, and P.A. Smith, “The Use of the PeakForce Quantitative Nanomechanical Mapping AFM-based Method for High-resolution Young’s Modulus Measurement of Polymers,” Meas. Sci. Technol. 22 (2011) 125703.
- G. Pletikapic, A. Berquand, T. Misic, and V. Svetlicic, “Quantitative Nanomechanical Mapping of Marine Diatom in Seawater Using Peak Force Tapping AFM,” J. Phycol. 48 (2012) 174–85.
- C. Heu, A. Berquand, C. Caille-Elie, and L. Nicold, “Glyphosate-induced Stiffening of HaCat Cells, a PeakForce Tapping Study on Live Cells,” J. Struct. Biol. 178 (2012) 1–7.
- I.N. Sneddon, “The Relation between Load and Penetration in the Axisymmetric Boussinesq Problem for a Punch of Arbitrary Profile,” Int. J. Eng. Sci. 3 (1965) 47–57.
- B. Luo, R. Yang, P. Ying, M. Awad, M. Choti, and R. Taylor, Proc. IEEE 32nd Annual Northeast Bioengineering Conference (IEEE, 2006), 81–82.
- D. Lobsien, A.Y. Dreyer, A. Stroh, J. Boltze, and K-T. Hoffmann, PloS One 8 (2013) e62644.
- R. Deepthi, R. Bhargavi, K. Jagadeesh, and M.S. Vijaya, SASTECH 9, (2010) 27–30.
- S. Pichardo, J. Kivinen, D. Melodelima, and L. Curiel, Physics in Medicine and Biology 58 (2013) 2163–83.
- V. Mirabet, P. Das, A. Boudaoud, and O. Hamant, “The Role of Mechanical Forces in Plant Morphogenesis,” Annu. Rev. Plant Biol. 62 (2011) 365–85.
- D.W. Ehrhardt and W.B. Frommer, “New Technologies for 21st Century Plant Science,” Plant Cell. 24 (2012) 374–94.
- P. Milani, S.A. Braybrook, and A. Boudaoud, "Shrinking the Hammer: Micromechanical Approaches to Morphogenesis," J. Exp. Botany 10 (2013) doi:10.1093/jxb/ert169.
- P. Milani, M. Gholamirad, J. Traas, A. Arnéodo, A. Boudaoud, F. Argoul, and O. Hamant, “Invivo Analysis of Local Wall Stiffness at the Shoot Apical Meristem in Arabidopsis Using Atomic Force Microscopy,” Plant J. 67 (2011) 1116–23.
- Q. Li, G.Y.H. Lee, C.N. Ong, and C.T. Lim, “AFM Indentation Study of Breast Cancer Cells,” Biochem., Biophys. Res. Commun. 374 (2008) 609–13.
- S. Morreno-Flores, R. Benitez, M. Vivanco, and J.L. Toca-Herrera, “Stress Relaxation and Creep on Living Cells with the Atomic Force Microscope: a Means to Calculate Elastic Moduli and Viscosities of Cell Components,” Nanotechnology 44 (2010) 445101–10.
- E.C. Faria, N. Ma, E. Gazi, P. Gardner, M. Brown, N.W. Clarke, and R.D. Snook, “Measurement of Elastic Mechanical Properties of Cancer Cells by Atomic Force Microscopy,“ Analyst 133 (2008) 1498–1500.
- A. Berquand, H.M. Kuhn, A. Holloschi, J. Mollenhauer, and P. Kioschis, “Expression of Tumor Suppressors PTEN and TP53 in Isogenic Glioblastoma U-251MG Cells Affects Cellular Mechanical Properties - An AFM-based Quantitative Investigation,” submitted to J. Appl. Phys (2013).
Authors
- Bede Pittenger, Andrea Slade, Alexandre Berquand (Bruker Nano Surfaces)
- Pascale Milani, Arezki Boudaoud, and Olivier Hamant (Ecole Normale Superieure de Lyon, France)
- Special thanks to Manfred Radmacher (University of Bremen, Germany)
©2013 Bruker Corporation. All rights reserved. BioScope Catalyst, MIRO, NanoScope, PeakForce Capture, PeakForce KPFM, PeakForce QNM, PeakForce Tapping, ScanAsyst, and TappingMode are trademarks of Bruker Corporation. All other trademarks are the property of their respective companies. AN141, Rev. A0