Instant and automated typing
Automated searching for the differentiating needle in the haystack
Dive into deeper microorganism characterization with the unique MBT HT Subtyping Module. In a single streamlined workflow, this innovative module automates the detection of specific marker peaks or peak profiles, harnessing the extraordinary discriminatory power of MALDI-TOF mass spectrometry. As thousands of microorganism species reveal their identity with unprecedented speed and accuracy, our powerful software seamlessly conducts further typing and result reporting, contributing to your microbial insights.
upon successful identification
for potential resistance
Elevating differentiation in microorganism identification
Bruker's MALDI Biotyper® has redefined microorganism identification, and the MBT HT Subtyping Module takes this legacy even further. While our MALDI Biotyper® excels in identifying diverse species, certain genera pose challenges in differentiation. The MBT HT Subtyping Module automates the detection of specific marker peaks, instilling newfound confidence in microbial identification.
Our team engineered the MBT HT Subtyping Module to surmount these challenges, automating the differentiation of complex cases such as Mycobacterium chimaera from Mycobacterium intracellulare, and species confirmation of Streptococcus pneumoniae, S. mitis_oralis and S. pseudopneumoniae.
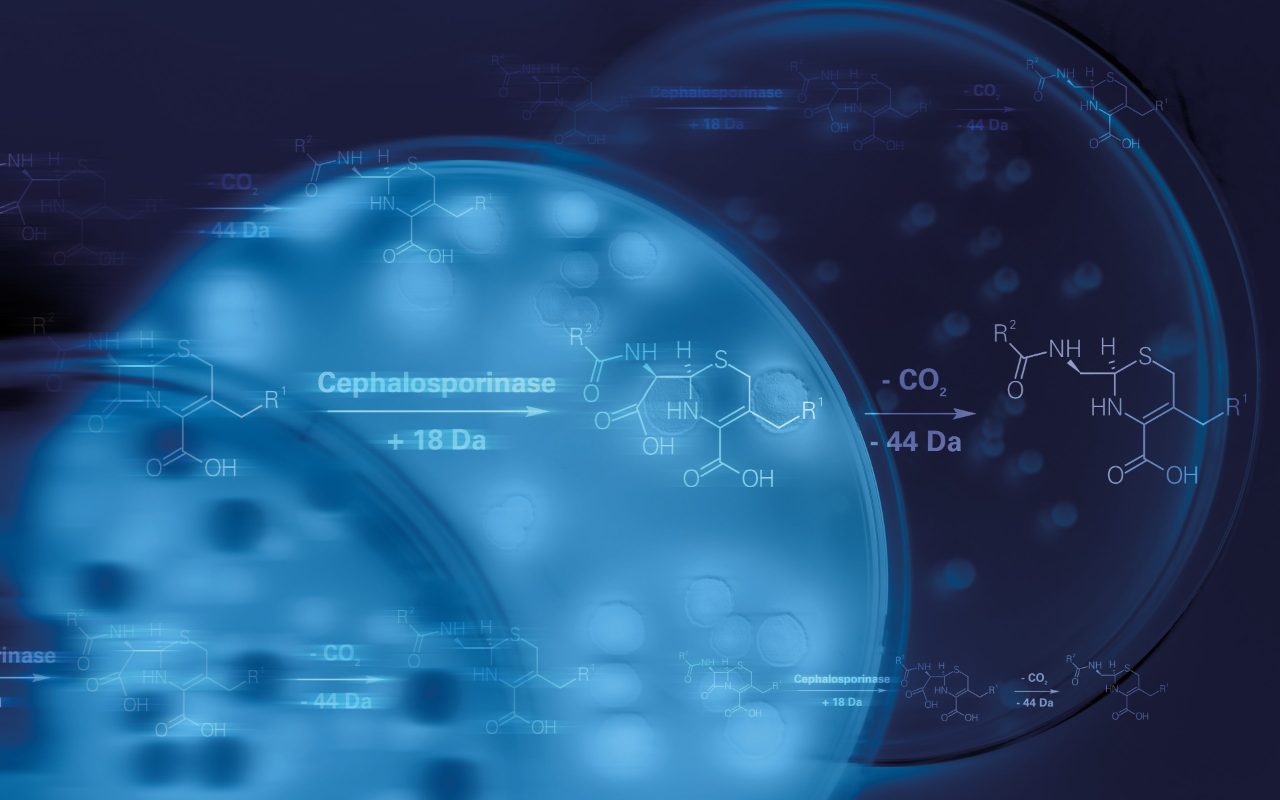
Empowering detection of resistance markers
The need for precise microorganism differentiation resonates across industries, from healthcare to food production, as we combat antibiotic resistance and track infectious outbreaks. Implement the MBT HT Subtyping Module, designed to swiftly pinpoint resistance markers tailored to these crucial applications.
As antibiotic-resistant bacteria surge, effective prevention and control are paramount. The potential of MALDI-TOF mass spectrometry to identify specific resistance markers is harnessed through the MBT HT Subtyping Module, sounding a warning bell for potential resistance. In a single automated workflow, it seamlessly merges pathogen identification with the automated detection of resistance markers, offering early warnings for blaKPC expressing Klebsiella pneumoniae and Escherichia coli, and Bacteroides fragilis cfiA positive strains.
Efficiency amplified: no additional sample preparation
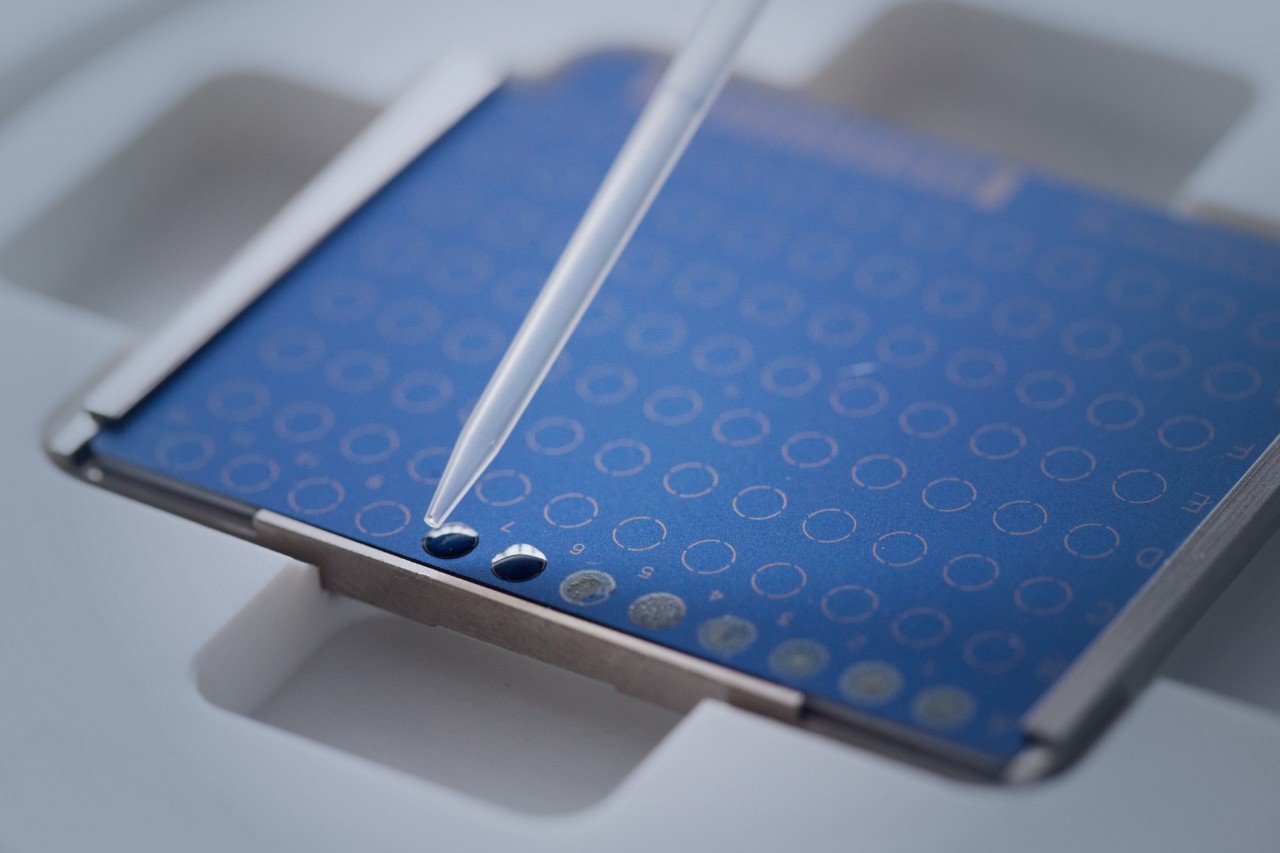
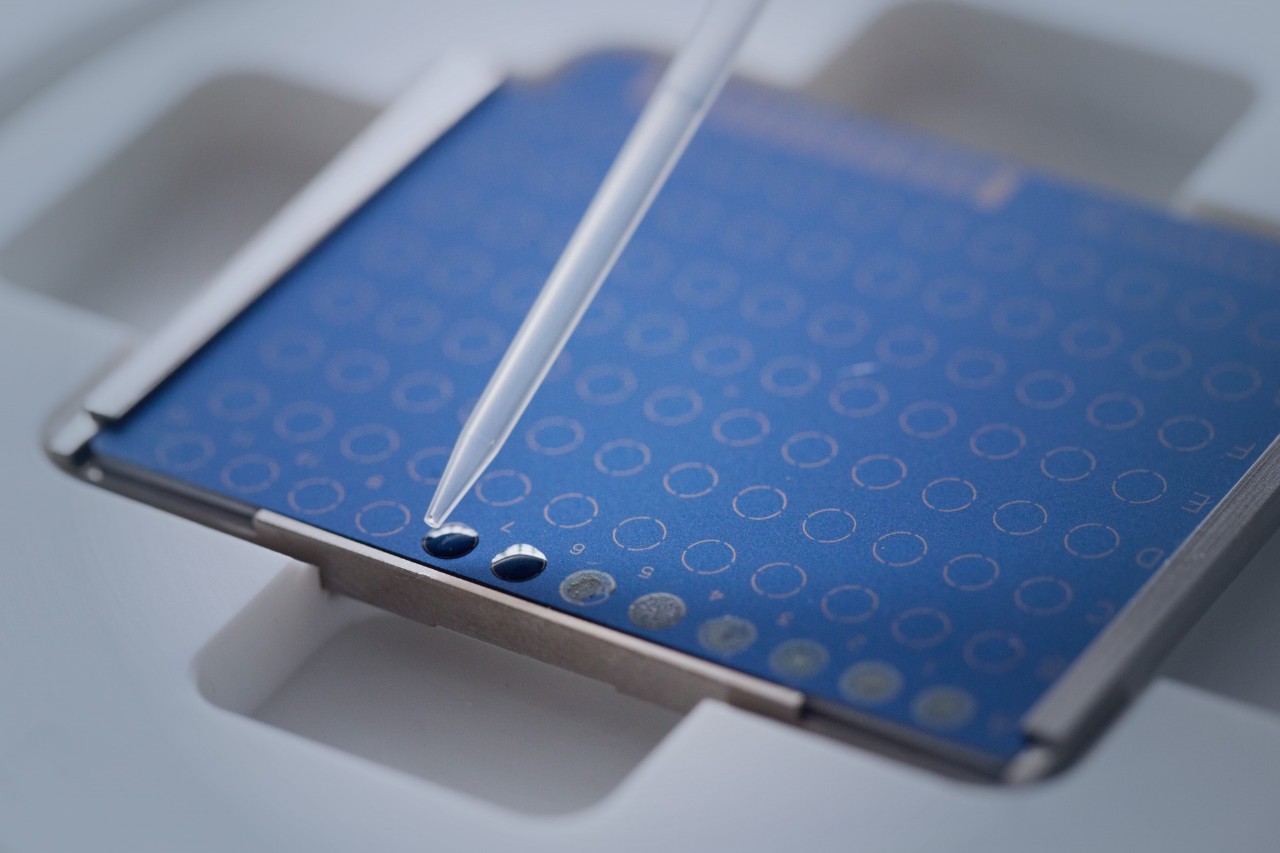
Elevate your microbial insights effortlessly. With the MBT HT Subtyping Module, no additional sample preparation is required once samples are placed on MALDI target plates. Upon microbial identification, our software takes charge, conducting deeper typing and result reporting automatically. This thorough process optimizes laboratory efficiency, ensuring swift time-to-result and propelling your research forward.
Subtyping solutions fitting to your microbiology lab
Bruker’s subtyping solution is available in two different versions. Please contact your local representative for availability in your country:
MBT HT Subtyping Module
For Research use only. Not for use in clinical diagnostic procedures.
MBT HT Subtyping IVD Module
For professional use only.
Not for sale in the USA or Puerto Rico.