Assembling Protein-Mediated DNA Bridges and Measuring Interaction Forces
Investigating Protein-Mediated DNA Bridges and Interaction Forces with Optical Tweezers
The NanoTracker Optical Tweezers (OT) setup delivers the ground-breaking capabilities researchers need to unravel fundamental processes taking place in living cells. This system enables the non-invasive trapping and manipulation of biological samples in both in vitro and in vivo studies. It is a powerful research tool for investigating real-time interactions in living cells, single molecules, viruses, and bacteria with nanometer precision and femtonewton resolution.
This application note outlines how optical tweezers can be successfully applied to study DNA damage, repair mechanisms, and protein-mediated DNA bridges. We demonstrate that interaction forces between individual molecules, the strength of the intermolecular connection, and the rupture force necessary to break protein-mediated DNA bridges apart can be precisely measured and quantified.
Readers can expect to learn about:
- The advantages of non-invasive trapping and manipulation of biological samples;
- How dual and multi-trap OT can be combined with fluorescence microscopy to assemble and visualize complex single-molecule assays and study their function in vitro; and
- How to use the NanoTracker to manipulate, trigger, and probe DNA-DNA, DNA-protein, and protein-protein interactions.
KEYWORDS: Optical Tweezers; NanoTracker; Fluorescence Microscopy; Optical Integration; Optical Trap; Force Measurements; DNA; DNA Bridges; Nanomanipulation
The importance of optical tweezers as a single-molecule technique cannot be overemphasized. A highly focused laser beam is used to trap and manipulate micrometer-sized objects with unprecedented precision and to measure molecular interaction forces. Dual- and multi-trap optical tweezers combined with fluorescence imaging methods enable the assembly and visualization of complex single-molecule assays and the study of their function in vitro. Over the past few decades, sophisticated optical tweezing experiments have provided valuable insights into fundamental processes, such as DNA repair, compaction, recombination, and transcription, etc.,[1-4] taking place in living cells where DNA molecules and related proteins play a central role.
DNA in cells is damaged continuously as a result of both internal and external factors. Various intracellular processes can lead to base modifications, such as alkylation and oxidation, or base pair mismatches during synthesis. External factors that cause DNA damage include free radicals, ultraviolet light ionizing radiation, and intercalation of small molecules between the DNA bases. Any DNA damage can lead to corruption of the genome, disrupt cell functionality, and potentially lead to the development of various diseases including immunological disorders and cancer.
Cells have several tailored mechanisms to repair DNA and an assortment of DNA-associated proteins and enzymes which scan the DNA for damage and initiate biochemical reactions to maintain and restore genome integrity.[5]
Role of XRCC4-XLF proteins in DNA repair
In human cells, the DNA-repair mechanism “classical Non-Homologous End Joining” (cNHEJ) is the main pathway for the repair of double-strand breaks (DSB), the most severe form of DNA damage. Once a DSB is detected, the ends of the damaged DNA are protected to prevent further degradation and damage. The two extremities of the broken DNA then need to be connected together for processing and to trigger the final ligation (see Figure 1). The cNHEJ repair process does not rely on a homologous template and, as such, must be very fast to be successful. In fact, unless every stage of cNHEJ is fast enough, the two extremities of the broken DNA will diffuse apart and correct ligation will no longer be possible.
XRCC4 (X-ray Repair Cross-Complementing Protein 4) and XLF (XRCC4-Like Factor) are two proteins known to participate in the cNHEJ DNA repair pathway. They can bridge broken DNA strands and hold them together. In solution, XRCC4 and XLF form filaments, consisting of a chain of alternating XRCC4 and XLF molecules, that bind to double-stranded DNA, allegedly by wrapping around it.
Triggering interactions between XRCC4-XLF-decorated DNA molecules
The NanoTracker 2 optical tweezers system enables the direct study of DNA-DNA interactions mediated by protein complexes. To characterize the interaction of XRCC4-XLF filaments with DNA molecules and their DNA- bridging mechanism, it is necessary to manipulate two DNA molecules simultaneously, requiring at least four optical traps be generated. The NanoTracker 2 Traps Multiplexing feature[7] enables the generation of multiple, stable optical traps from a single laser source in a time-shared manner with precise control of the trap position in three dimensions (see Figure 2A). It is therefore possible, for example, to catch two DNA molecules with biotinfunctionalized ends from laminar flow in a multichannel microfluidics chamber using streptavidin-coated polystyrene beads (5 µm in diameter). By holding and changing the position of the trapped beads, it is then possible to manipulate, trigger, and probe DNA-DNA, DNA-protein, and protein-protein interactions in different configurations.[8,9] In the case of protein studies, the proteins of interest can be pre-incubated with the DNA or injected at will through a different channel of the above mentioned microfluidics device.
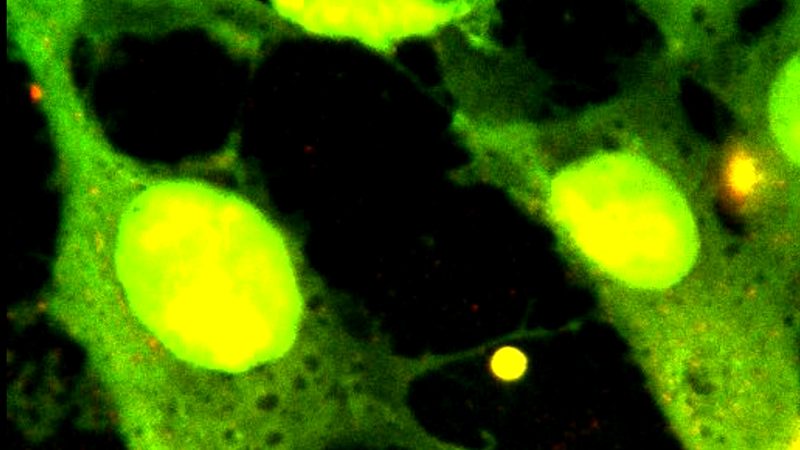
Furthermore, single-molecule fluorescence imaging can be performed simultaneously to tweezing with the NanoTracker 2 system. This enables the direct detection and observation of fluorescently tagged molecules in real time and the quantitative analysis of the fluorescence intensity. Figure 2B shows an epi-fluorescence image of two DNA molecules stained with the dye Sytox Orange, acquired with the NanoTracker 2 equipped with an EMCCD camera (iXonEM+, Andor).
For prolonged experiments, it is not recommended to fluorescently label the DNA molecules themselves, as even low photodamage intercalating dyes have been shown to affect the properties and structure of DNA[10-12] and continued imaging can lead to DNA breakage. A better approach is to use fluorescently labeled proteins instead, which dramatically reduces photodamage and permits the direct observation and localization of the proteins being studied on the DNA molecules. Figure 2C shows a single frame of eGFP-labeled XRCC4-XLF complexes decorating two DNA molecules. The DNA molecules are being held between four trapped beads and are brought into contact by accurate positioning of the traps. At the DNA-DNA contact point, a very bright fluorescent spot can be observed, indicating an accumulation of XRCC4-XLF proteins. Furthermore, the XRCC4- XLF-mediated bridge is shown to effectively withstand the forces being applied to pull the traps apart.
The multiple traps generated by the NanoTracker 2 can be moved independently in three dimensions within the sample and different geometries can be used to study the interaction between DNA molecules. Figure 3 shows three different ways to manipulate a pair of DNA molecules decorated with XRCC4-XLF by bringing them into contact using different configurations.
Figure 2.
A: Complex arrangements of multiple DNA molecules for studying DNA-protein interactions can only be implemented with several independentlycontrolled traps.
B: Fluorescence image of two DNA molecules held between four polystyrene beads. DNA is stained with the dye Sytox Orange.
C: Fluorescence image of XRCC4-XLF proteins bridging two DNA molecules (blue arrow). XLF is labelled with eGFP. Scale bars in B and C are 5 µm.
Figure 3.
Different configurations used to bring two DNA molecules in contact and form XRCC4- XLF-mediated DNA bridges. Each image shows a selected frame, representing a characteristic stage of each manipulation, from a fluorescence acquisition where XLF was labelled with eGFP.
A: (“Wrappingunwrapping” procedure): One of the four trapped beads is manipulated in 3D to wrap one DNA molecule around another one.
B: (“Pairing”) and
C: (“Aligning”) procedures: DNA molecules are brought in contact by combining the manipulation of bead positions and the use of shear forces (blue arrows show flow direction). Solid and dashed green circles indicate the current and previous position of the bead being moved, respectively. The 10 µm scale bars are allocated for all image panels (A, B and C).
For example, one DNA molecule can be wrapped around another, later unwrapped, and subsequently probed to see whether the two DNA molecules have been effectively bridged by the XRCC4-XLF protein complex. A series of sequential images (see Figure 3A) show the “wrapping” procedure in detail. This procedure begins with one DNA molecule being held in a static position while the second DNA is brought in contact by manipulating the position of the trapped beads. Next, one end of the second DNA molecule is moved vertically in the Z direction, which is possible thanks to NanoTracker 2’s axial control of the trapping position. Next, the second DNA is moved over the static DNA molecule and then downwards until it is below the static DNA, again using axial positioning. The second DNA molecule is then brought underneath the static molecule, and finally moved up again until it is at the original axial position, thereby completing a full wrap of one DNA around the other one. Once the two DNA molecules and XRCC4-XLF complex have been left to interact together for a specified amount of time, the molecules are unwrapped by performing the manipulation steps described above in the reverse order. After being unwound, the two DNA molecules remain connected as a result of the XRCC4-XLF-mediated bridging.
Another approach to trigger XRCC4-XLF-mediated interactions between DNA molecules, called “Pairing”, is shown in Figure 3B. In this configuration, one DNA molecule is again straightened and kept static. The second DNA is brought in contact with the static DNA using the microfluidic flow to stretch it, and by moving the attached beads towards the static DNA until the two molecules touch. Similar to the previous example, use of the “pairing” configuration enables control of the interaction time between the molecules. Finally, the beads are separated and the established XRCC4-XLF-mediated bridge can be tested.
Figure 3C shows a further possible configuration, called “Aligning”, to generate DNA-DNA interactions. In this case, only two traps are necessary to bring the DNA molecules in contact with each other. Each DNA molecule is attached to a trapped bead at one end and the other end is free. By applying a moderate buffer flow, the molecules are straightened and aligned along the flow as a result of shear forces between the DNA and the flowing buffer. The trapped beads are then carefully manipulated until the DNA molecules are brought into contact with each other, enabling XRCC4-XLF to interact with and bridge the two DNA molecules. The beads are then separated in a controlled manner and the connection between the DNA molecules is probed.
Measuring protein-mediated DNA bridging forces
One of the most remarkable capabilities of modern optical tweezers setups, such as the NanoTracker 2, is the ability to measure interaction forces between individual molecules. Several procedures have been described above to create connections between DNA molecules decorated with bridging proteins using XRCC4-XLF as an example. A similar approach can be used for any other protein, protein complex, or mediators of interactions between DNA molecules. Qualitative information on how molecules interact and how proteins are distributed along the DNA can be obtained by performing fluorescence imaging while using optical tweezers to apply forces and manipulate the DNAs-proteins complex. Furthermore, once a DNA bridge has been formed via the steps described above, it is possible to measure the strength of the intermolecular connection established. As the NanoTracker 2 can measure forces precisely, it is ideal for quantifying the rupture force necessary to break apart protein-mediated DNA bridges.
Once a DNA bridge has been created, the DNA molecules should be aligned by arranging the positions of the trapped beads along the X and Y-axis in a cross-like configuration (see Figure 4).
Then, one bead should be pulled by the optical trap, for example, along the X-axis to exert a force on the DNA-protein-DNA complex, while the other traps are kept static. The trap opposite the moving one is used to measure the force exerted along the X-axis. Assuming the forces acting along the Y-axis are negligible (f1 ~ f2 ~ 0), the peak (maximum) force F measured during the force-spectroscopy experiment represents the rupture force necessary to break the bridge apart.
Repeating this rupture force measurement for a certain number of connections provides a statistically significant distribution of the bridging force required to break apart a specific protein-mediated DNA-DNA bridge. Figure 5, for example, shows the cumulative probability distribution of the rupture force measured for XRCC4-XLF-mediated connections. These data provide insight into the range of forces that XRCC4-XLF-mediated DNA bridges can withstand, and can be used to compare and gauge different proteinmediated DNA bridges. As can be seen in the cumulative probability distribution plot in Figure 5, the rupture forces of XRCC4-XLF-mediated DNA bridges lie roughly between 5 pN and 80 pN, and more than 50 % of the rupture forces measured were higher than 40 pN.
Conclusion
The NanoTracker 2 is a powerful optical tweezers setup that enables manipulation of objects with nanometer accuracy and measurement of molecular interactions with piconewton resolution. Here, we have detailed several different procedures that outline how optical tweezing can be successfully applied to assembling protein-mediated DNA bridges. The current toolbox can be directly applied for in vitro reconstitution studies to study the fundamental processes taking place in living cells.
Authors
- Vitaliy Oliynyk, Ph.D., JPK BioAFM, Bruker, Berlin (Germany)
- Davide Normanno, Ph.D., Cell Biology of RNA & Chromatin and Cell Biology Labs, Institute of Human Genetics, Univ Montpellier, CNRS, Montpellier (France).
References
- Heller, I., Hoekstra, T. P., King, G. A., Peterman, E. J., & Wuite, G. J. (2014). Optical tweezers analysis of DNA–protein complexes. Chemical reviews, 114(6), 3087-3119.
- Mukhortava, A., Pöge, M., Grieb, M. S., Nivina, A., Loot, C., Mazel, D., & Schlierf, M. (2019). Structural heterogeneity of attC integron recombination sites revealed by optical tweezers. Nucleic acids research, 47(4), 1861-1870.
- Gibbs, D. R., Mahmoud, R., Kaur, A., & Dhakal, S. (2021). Direct unfolding of RuvA-HJ complex at the single-molecule level. Biophysical Journal, 120(10), 1894-1902.
- Brouwer, I., Sitters, G., Candelli, A., Heerema, S. J., Heller, I., Zhang, H., Normanno, D., Modesti, M., Peterman, E., & Wuite, G. J. (2016). Sliding sleeves of XRCC4–XLF bridge DNA and connect fragments of broken DNA. Nature, 535(7613), 566-569.
- Ceccaldi, R., Rondinelli, B., & D’Andrea, A. D. (2016). Repair pathway choices and consequences at the double-strand break. Trends in cell biology, 26(1), 52-64.
- Lans, H., Marteijn, J. A., & Vermeulen, W. (2012). ATP-dependent chromatin remodeling in the DNA-damage response. Epigenetics & chromatin, 5(1), 1-14.
- JPK BioAFM by Bruker, (De-)Multiplexing: multi-trap force measurements and sample manipulation with JPK’s NanoTracker 2, Technical Note. https://www.jpk.com/apptechnotes-img/Optical-Tweezers/pdf/jpk-tech-nt-de-multiplexing.1.pdf
- JPK BioAFM by Bruker, Single-molecule DNA (over)stretching using optical tweezers, Application Note. https://www.jpk.com/app-technotes-img/Optical-Tweezers/pdf/jpkapp-nt-dna-stretching.1.pdf
- JPK BioAFM by Bruker, Force clamping in arbitrary directions, Technical Note. https:// www.jpk.com/app-technotes-img/Optical-Tweezers/pdf/jpk-tech-force-clamping.1.pdf
- Thakur, S., Cattoni, D. I., & Nöllmann, M. (2015). The fluorescence properties and binding mechanism of SYTOX green, a bright, low photo-damage DNA intercalating agent. European Biophysics Journal, 44(5), 337-348.
- Tycon, M. A., Dial, C. F., Faison, K., Melvin, W., & Fecko, C. J. (2012). Quantification of dye-mediated photodamage during single-molecule DNA imaging. Analytical biochemistry, 426(1), 13-21.
- Biebricher, A. S., Heller, I., Roijmans, R. F., Hoekstra, T. P., Peterman, E. J., & Wuite, G. J. (2015). The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nature communications, 6(1), 1-12.
©2022 Bruker Corporation/Bruker Nano GmbH. All other trademarks are the property of their respective companies. All rights reserved. AN304 Rev. A0.