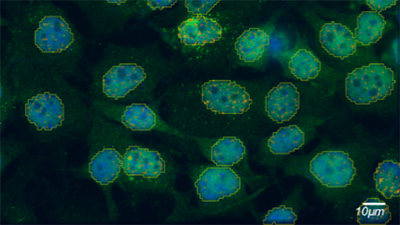
Tech Note: Label Selection and Strategies for Super-Resolution Single Particle Tracking
Successfully visualize dynamic biological processes
This technical note discusses how to choose a single particle tracking (SPT) labeling strategy based on your specific experimental design and requirements.
Contents include:
- An overview of SPT labeling strategies, including genetically encoded labels, non-genetically encoded labels, and live tracking of RNAs
- Explanation of the advantages of each strategy
- Notes about these strategies' compatibility with common experimental conditions and imaging techniques
- Practical considerations and tips for choosing the best genetic and chemical labeling techniques for your experiments
KEYWORDS: Single particle tracking, super-resolution microscopy, fluorescent proteins, labeling strategies, Vutara VXL, SRX software
Single particle tracking is a localization microscopy technique that allows researchers to visualize dynamic biological processes. There are several factors one must consider to ensure success, such as laser power, fluorophores, and experimental design. This technical note discusses several labeling strategies, including genetically encoded labels, non-genetically encoded labels, and live tracking of RNAs, as well as their advantages for various experimental requirements.
General Considerations for Single Particle Tracking
Single particle tracking requires the detection and localization of spots over time. Localization precision is eventually proportional to the square root of the number of photons of signal collected per spot, and brighter spots can be localized with greater precision. Therefore, brighter spots improve the quality of single particle tracks. For single particle tracking, brightness matters even more than for other localization techniques, as both spatial and temporal resolution are important.
For fast-moving particles or rapidly evolving processes, very high frame rates may be required to avoid blurring, meaning shorter windows to collect photons. In these situations, spot brightness becomes more important, and increasing excitation intensity can increase the number of photons collected per spot. Bruker’s Vutara VXL super-resolution microscope has watt-scale lasers for most imaging lines, allowing for very intense, even illumination across the entire field of view. However, high intensity illumination can lead to rapid spot bleaching, requiring a balance between intensity and duration for single particle tracking. Due to Vutara VXL’s suite of neutral density filters, stable illumination intensities in the range of milliwatts to kilowatts per square centimeter are possible on the same instrument with the same laser line, and if desired, during the same experiment.
LEARN MORE:
General Tips for Choosing the Correct Labeling Approach
With the exception of some specific tracking techniques discussed later, highly photostable fluorophores are desired. Ideally, these fluorophores should not bleach, blink, or photoconvert. This is in contrast to other single-molecule localization microscopy (SMLM) techniques, where blinking and photoconversion are desired. The brightest, most photostable label available, in most cases, is quantum dots. However, getting them attached to the structure or material of interest in the correct context can be difficult, as they must be introduced into the cell already attached to the protein/object of interest, linked to an antibody, or directly ligated through click chemistry. Quantum dots, particularly when attached to an antibody, are too large to freely pass through the membrane. This requires introduction into the cell via microinjection, transfection, or other disruptive approaches when targeted against intracellular components.
Another consideration is ease of use. Genetic encoding for labels can be very helpful in some circumstances and can simplify the process of sample labeling, since it’s oftentimes easier to transfect a plasmid than to make and inject quantum dot–labeled proteins. However, genetic labels can come with their own drawbacks, such as needing sparse labeling (only a few particles per cell) to track. Sparse labeling can be achieved, among other methods, with endogenous tagging of sparse structures (e.g., centrosome, or specific mRNAs) or low-expressing, low-copy-number plasmids.
SPT-PALM (single-particle tracking photoactivated localization microscopy), provides another option for sparse labeling by photoconversion or photoactivation of a subset of dyes by brief pulses of photoactivation, followed by longer-duration recording and tracking.
Selecting the best approach and label for experiments can be a difficult decision. Our Bruker applications team is also happy to provide assistance with specific experimental questions. Furthermore, Vutara VXL instruments are compatible with all methods discussed, and its powerful SRX software can image and track particles, whether they are quantum dots, fluorescent proteins, or individual dye molecules.
Genetically Encoded Labels
Fluorescent Protein (FP) selection is important for single particle tracking. Key factors in selection should be brightness and photostability. New fluorescent proteins are being developed all the time, so this list will be out of date as soon as it’s published, but here are a few suggestions from the applications team at Bruker. When choosing an FP for a new line, strain, or plasmid, consider checking out FPbase, or, for more assistance, contact the Bruker applications team. Furthermore, keep in mind that to track stably fluorescent proteins, the labeled structure must be naturally sparse, with only a few spots present per cell at a time.
Halo and SNAP-Tags
Halo or SNAP -tagging proteins provides the flexibility to label now and choose the wavelength and technique later. Additionally, a wide array of specialized chemical dyes is available, which may provide additional benefits, including Halo-functionalized self-blinking dyes, and photo-activatable dyes for PALM and SPT-PALM. Halo and SNAP-tagged cell lines are commercially available, and strains/lines of model organisms may already exist for a given protein of interest.
Photoconvertible fluorescent proteins are also usable for SPT-PALM. Photoconvertible FPs are typically less bright and have worse photoconversion kinetics than chemical dyes. However, cell, tissue, and animal lines/strains may exist for different applications, as photoconvertible fluorescent proteins have been used extensively for time marking and flexibility in developmental and cell biology. For SPT-PALM, it is generally recommended to use HALO- or SNAP-tagged proteins and PA-JF dyes for visualization.
Non-Genetically Encoded Labels
Sometimes it will be possible to label a protein, RNA, biomolecule, or structure of interest directly or indirectly with a brightly fluorescent object or dye and inject, transfect, or electroporate it into the sample for subsequent study. This is an excellent, albeit low-throughput and potentially technically complex, approach for single-particle dynamics studies. Consider using:
Quantum Dots
- Highly fluorescent inorganic structures
- Available functionalized
Gold Nanourchins
- Inert spiky gold particles (emit light via surface plasmon resonance rather than fluorescence, and do not bleach)
- Available functionalized
Primary Antibodies or Nanobodies
- Directly labeled primary or secondary antibodies against the biomolecule of interest (if the structure is on the outside of the cell at the beginning of the experiment, e.g., receptor extracellular domains, this may be a viable strategy)
- Labeled primary antibodies and labeling kits are commercially available
LEARN MORE:
Live Tracking of RNAs
For live tracking of RNAs, there are several techniques, each of which will likely work with the Vutara VXL.
- MS2/MCP (and similar techniques)
- Peppers (and similar techniques)
- Transfecting pre-labeled RNAs
For detailed advice on selecting methods and labels for RNA imaging, please contact the applications team.
Single Particle Tracking with Vutara VXL
Successful single particle tracking relies on thoughtful label selection tailored to specific biological questions. Each strategy offers unique advantages and trade-offs. Whether leveraging the brightness and stability of quantum dots, the genetic precision of fluorescent proteins, or the flexibility of Halo and SNAP-tags, a number of factors need to be considered and balanced against experimental goals. These factors include photostability, labeling density, and ease of implementation. The unique capabilities of the Vutara VXL super-resolution microscope and SRX software make this entire process easier and more precise for advanced exploration of dynamic molecular processes.