Walking Along Chromosomes: Visualizing in situ Chromosomal Structure with Single Molecule Localization Microscopy
Discover what SMLM can bring to your genetics research
During this webinar, guest speakers Dr. Chao-ting Wu and Dr. Guy Nir will discuss their research using SMLM and OligoStorm to investigate chromosomal structures in situ. Topics discussed will include:
- Single molecule localization microscopy and OligoSTORM to image chromosomes
- Sequential fluorescent labeling to image complex molecules
- Interpreting 3D images of chromosomal structure
- Functional implications of the 3D structure of chromosomes
Webinar Summary
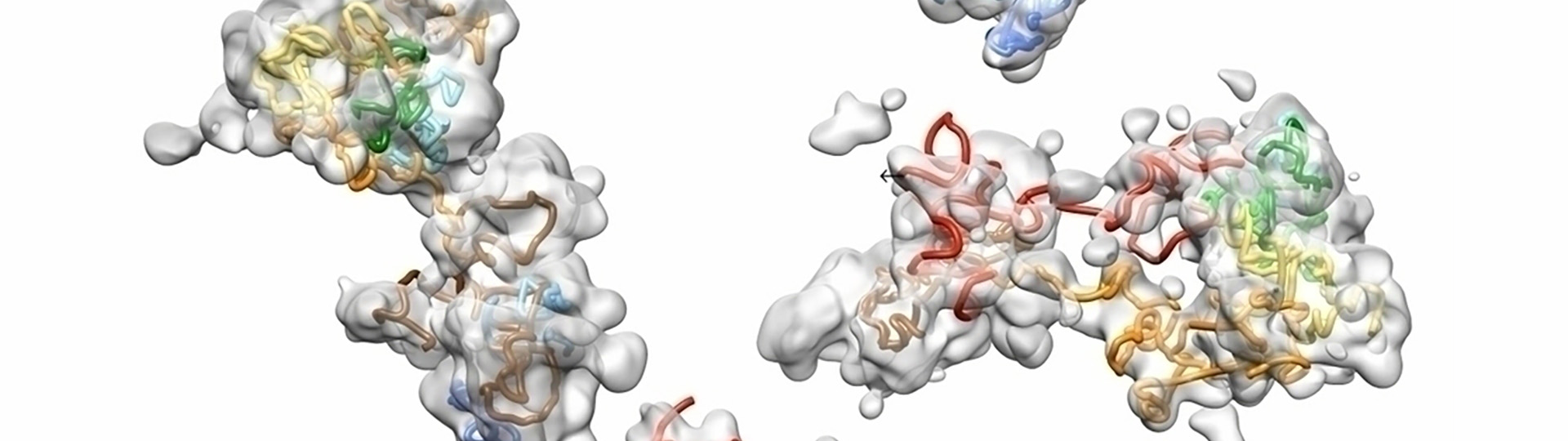
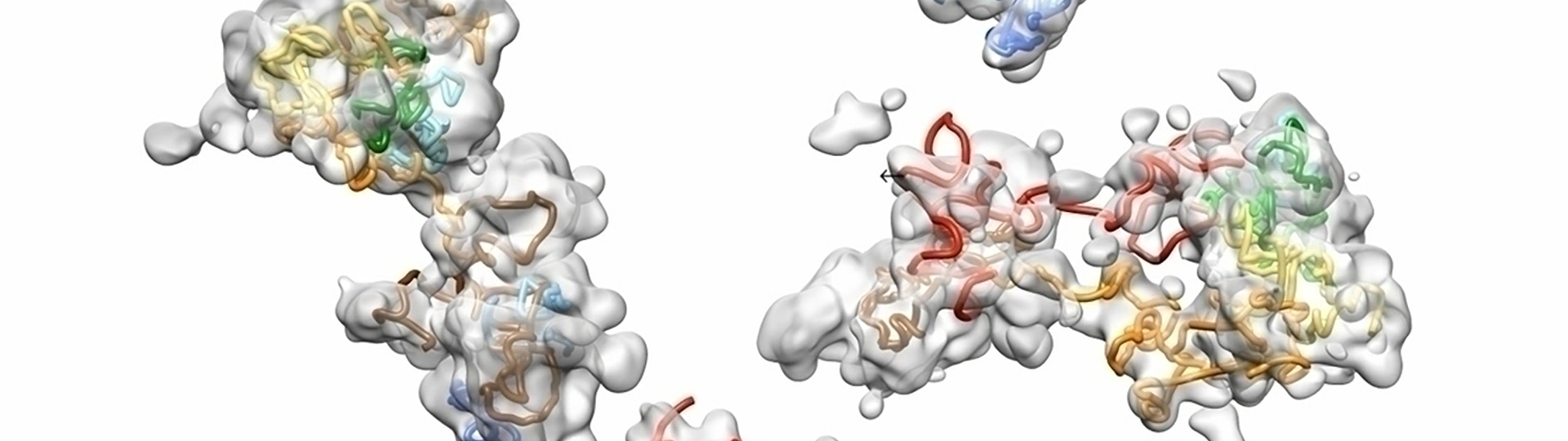
Understanding chromosomal structure in situ is critical to understanding nuclear function. This webinar shows how OligoSTORM and single molecule localization microscopy can be used to visualize in situ chromosome structure. Microfluidics and sequential labeling strategies can be employed to “walk” along chromosomes to visualize chromosome regions that form distinct structures in the form of compartments.
Data presented, up to 8 megabases of human chromosome 19, will show that there may be differential organization of chromosomes in maternal and paternal homologous in addition to regional compartments. We will also discuss the functional implications of these findings and the types of data that can be generated with these techniques in terms of understanding chromosomal function.
Find out more about our solutions for Single Molecule Microscopy or our other solutions for Super-Resolution Microscopy:
Featured Products and Technology
Speakers
Dr. Chao-ting Wu
Professor, Department of Genetics, Harvard University
Dr. Guy Nir
Post-doctoral Fellow, Department of Genetics, Harvard University


